From the journals: MCP
Metals contaminate protein samples. Lipid droplets go haywire in Huntington’s. Nerve cells respond to stimulation. Find out the latest from the journal Molecular & Cellular Proteomics.
Remove contaminating metal to study activated proteins
The bad news: Unwanted metal ions are knocking useful tags off proteins — tags that indicate that a protein is overactive in, say, cancer or metabolism. The good news: It’s possible to filter that metal out.
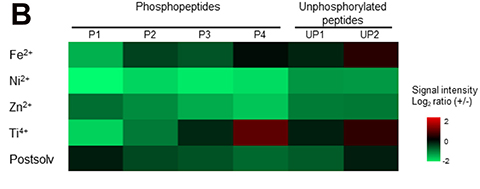
Get rid of metals from the mobile phase if you’re studying protein activation. That’s the conclusion of new research published in the journal Molecular & Cellular Proteomics. Removing metal contamination with a simple device yielded at least 10 times more tagged protein fragments than a traditional setup.
Proteins active in health or disease often have a phosphate tag attached (or two, or three). Researchers may catalog these tagged proteins at scale using phosphoproteomics. The first step is separating proteins in a machine: high-performance liquid chromatography. But there’s a catch: Metal in that equipment tends to contaminate samples, knocking tags off proteins — while at the same time, metal enables the lines and valves of the equipment to withstand high pressure. Some scientists have made metal-free components, but they don’t work for trace samples, which require thinner parts. And, until this study, the removal of metal ions hadn’t been tested for nanoscale samples, where narrow lines and extra contact time mean even more metal ions in the mobile phase (that’s the liquid protein samples are dissolved in).
Yumi Komori and a team of researchers at Kyoto University used a device to filter out metal ions from the mobile phase. Compared with unfiltered samples, they measured a 10-fold recovery of protein fragments attached to a single phosphate tag. For protein fragments with more than one tag, recovery increased 77 times versus controls. The team suggested that the technique may improve the recovery of samples not only at nanoscale but also at microscale.
Lipid droplets go haywire in Huntington’s
Medium spicy noodles. Sorry, medium spiny neurons. The full set of proteins in medium spiny neurons — brain cells vulnerable to Huntington’s disease — has been published in the journal Molecular & Cellular Proteomics. The study confirms existing therapeutic targets and reveals new ones: particularly, the importance of proteins that process fats.
Huntington’s disease causes neurons to break down, and it tends to get worse over time. To pinpoint cellular processes going awry, Kizito-Tshitoko Tshilenge and a collaborative team from the United States compared protein populations in brain cells impacted by the inherited disease with those of healthy cells.
First, scientists read between the lines: Diseased cells had more activity in the scaffolding between cells known as the extracellular matrix than healthy cells did. Moving to the nucleus, critical errors would accumulate because cells affected by Huntington’s were less able to repair breaks affecting both sides of DNA: double-sized breaks. And, the study showed, diseased cells took in more lipids and degraded fewer than their healthy counterparts. It turns out medium spiny neurons — like medium spicy noodles, perhaps — can be fatty.
Contributing to the disease were changes in the amounts of several key proteins. Apolipoprotein E, a protein that helps cells process lipid droplets: Cells with Huntington’s disease had less of it. Minichromosome maintenance proteins, so named because they maintain DNA when it’s damaged: There were fewer of them, too. And a family of proteins whose job it is to maintain brain health, septins: These proteins were dysregulated, as is the case in Alzheimer’s disease and Parkinson’s disease.
How stimulation impacts brain cells
A team of scientists has investigated how different parts of brain cells in rats respond to stimulation: in this case, water deprivation. The study contributes to research on brain plasticity by documenting which proteins are where and which proteins jump from standby into action.
Cells in the study hailed from the hypothalamus: a part of the brain behind hormones, heart rate and hunger. In a model system shared in the journal Molecular & Cellular Proteomics, Soledad Bárez-López, André S. Mecawi and a team of scientists in Brazil and Bristol determined which proteins were active at the axon terminals (the parts of brain cells making contact with the nearby pituitary gland) versus those in the cell’s skeleton (called the cytoskeleton and made of pure protein strings rather than bone).
The results reflected the expected functions of different cell regions: Cell bodies expanded their cytoskeleton, while the ends of cells concentrated signaling to nearby cells nearby. (Axon terminals are very social.)
What was unexpected: In stimulated cells, proteins that would go on to become hormones became hyperphosphorylated, ending up with more than one phosphate tag. Adding extra phosphate tags might be a key step in how cells get hormones to be ready for release. Think of it as a pep talk. (These are peptides we’re talking about.)
Read the full paper at mcponline.org to find out which proteins came up most often in the research — including vasopressin, which controls how wide blood vessels are, and oxytocin, known as the love hormone.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

ASBMB announces 2026 JBC/Tabor awardees
The seven awardees are first authors of outstanding papers published in 2025 in the Journal of Biological Chemistry.

