How scientists identified a new neuromuscular disease
Accurate diagnosis is the first step toward successful treatment of any disease.
Early in the COVID-19 pandemic, the omicron variant was often mistaken for the common cold due to overlapping symptoms like sore throat and congestion. But while a cold is typically harmless, SARS-CoV-2 can be deadly.
Neuromuscular diseases such as muscular dystrophy and amyotrophic lateral sclerosis, or ALS, often share symptoms like muscle weakness, atrophy and difficulty walking or breathing. However, treatment depends on the underlying cause. For example, patients with ALS may be prescribed Riluzole to extend survival, while those with congenital myasthenic syndrome may take Pyridostigmine to improve muscle strength.
For degenerative disorders, early diagnosis can be the difference between effective treatment and irreversible decline. But what happens when, even after exhaustive testing, no diagnosis emerges?
Undiagnosed, not unheard

When symptoms remain unexplained, clinicians often refer patients to programs like the National Institutes of Health Undiagnosed Diseases Program, or NIH UDP. Researchers such as NIH UDP translational research scientist Marie Morimoto and associate investigator May Christine V. Malicdan combine clinical data with genetic analysis to crack diagnostic puzzles.

Malicdan compares complex disorders to a “sophisticated recipe,” where symptoms and genetic variants are ingredients. Understanding the disease requires dissecting each component individually.
“You’re understanding the main component now and later on you will be able to understand how it contributes to more complex mechanisms and genetics,” said Malicdan.
In a recent study, nearly 30 international researchers — including Morimoto and Malicdan — investigated nine individuals from eight unrelated families who showed similar neuromuscular symptoms. These included poor motor coordination, respiratory weakness, difficulty walking and speaking and low body weight and stature. Symptoms varied by patient, but all pointed to a multisystemic, neurological disorder.
Given the neurological nature of the symptoms, researchers conducted MRIs. Adult patients showed cerebellar atrophy, while infants did not, suggesting a progressive condition.
Invisible markers
To build a diagnostic profile, the team cataloged each patient’s physical symptoms and genetic variants. Each participant and their unaffected family members underwent genome or exome sequencing. Genome sequencing analyzes the full DNA sequence, while exome sequencing focuses on protein-coding regions.
All affected individuals carried biallelic variants in the RFC4 gene, meaning they inherited a defective copy from each parent. In one patient, a spontaneous mutation paired with a single inherited variant.
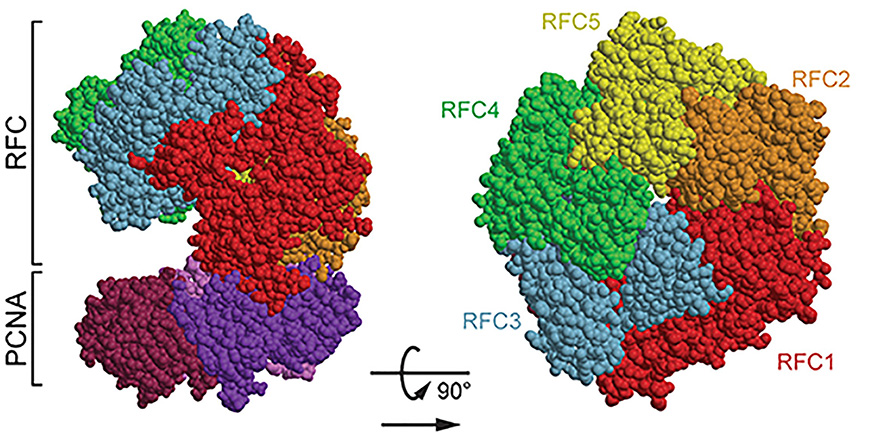
RFC4 encodes the protein replication factor C 4, or RFC4, one subunit of the replication factor C, or RFC, complex, which plays a key role in DNA replication and repair. Structural modeling and previous cryogenic electron microscopy data revealed that certain missense mutations, such as — p.Cys281Arg and p.Glu344Lys — likely disrupt RFC complex interactions.
In fibroblast cultures from affected patients, the RFC complex formed less efficiently, supporting the predicted impact of these mutations.
Disease: diagnosed
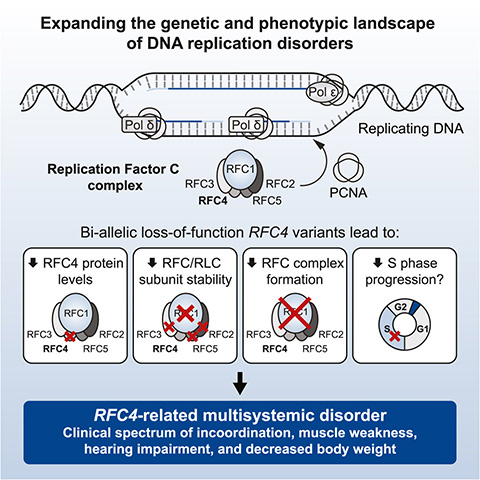
The condition is now recognized as Morimoto–Ryu–Malicdan neuromuscular syndrome, or MRMNS, an autosomal recessive disorder linked to RFC4 variants. The clinical and genetic data collected from the nine participants now form the basis for diagnosing future patients.
The collected clinical features and genetic data of the nine participants are now associated with MRMNS. Treatments can now be developed for MRMNS and any patient expressing similar symptoms can be quickly and correctly diagnosed.
Finding nine people with similar clinical presentations and finding a diagnosis that joins their presentations together is another day on the job for rare disease researchers. Each discovery adds to our broader understanding of disease origins and mechanisms.
The NIH UDP’s mission is to identify the genetic causes of rare disorders like MRMNS—not to focus solely on mechanistic deep dives. Morimoto and her colleagues want to “help as many patients as possible.”
Morimoto added that the publication has already helped others identify new cases.
“Our impact is finding those initial gene–disease associations to help other clinicians and other groups diagnose their own undiagnosed patients,” said Morimoto.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

