Unraveling the language of histones
Histones package DNA inside the nucleus, keeping it compact yet accessible. Their flexible tails act like signaling antennas, undergoing posttranslational modifications, or PTMs, that control DNA accessibility and gene activity. When these modifications go awry, the consequences can be serious, including cancer and other diseases.

Scientists like Philip Cole, a biochemist at Harvard Medical School, are uncovering how histone modifications influence gene expression and how correcting them could lead to new therapies.
Cole recently shared his research on the American Society for Biochemistry and Molecular Biology webinar Breakthroughs, a series highlighting research from ASBMB journals. A professor of medicine, biological chemistry and molecular pharmacology at Harvard Medical School, he also serves as an associate editor of the Journal of Biological Chemistry. During his talk, sponsored by JBC, Cole described how a therapeutic compound might correct dysregulated histone modifications and new chemical tools his lab developed to explore how these modifications interact.
Blocking histone modifiers
Histone tails are modified in many ways by enzymes such as histone deacetylases, or HDACs, which remove acetyl groups. HDACs often join forces with other enzymes to form multiprotein complexes. Cole studies one of these, the CoREST complex, short for corepressor for element-1 silencing transcription factor. This complex compacts DNA, effectively silencing genes by making them inaccessible for transcription.
“This complex has been implicated in many biological processes related to cancer … which has made this complex a potential therapeutic target for various cancers,” Cole said.
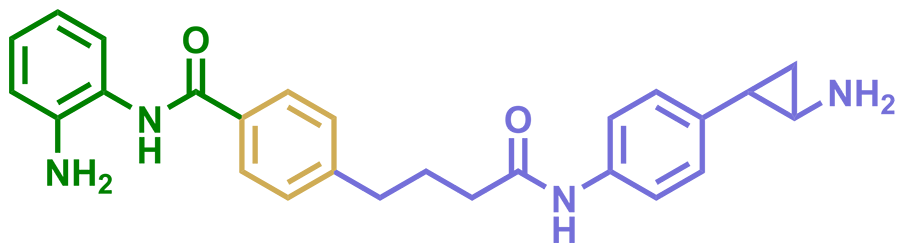
To understand the role of the CoREST complex in gene expression and disease, the team developed a selective small molecule inhibitor called corin to target its function. Using an in vitro assay, they observed reduced CoREST activity in the presence of corin. The team then used transcriptomics to investigate the effect of corin on melanoma cells and found that treated cells showed increased expression of tumor suppressor genes. Investigation of the therapeutic potential and molecular mechanisms of corin in cancers — including melanoma, breast and colon — is ongoing by Cole and collaborators.
Identifying PTMs and how they “talk”
Cole also works to understand the patterns and interactions of histone tail PTMs and focuses on a particular histone that is commonly modified, called H3. Yet, chemical tools to look at what PTMs are present and how they talk to each other were lacking. Proteomics approaches could be used to study PTMs, but the available methods weren’t ideal for histones because of where and how they cut up the proteins.
“This motivated us to try and improve upon this method,” Cole said.
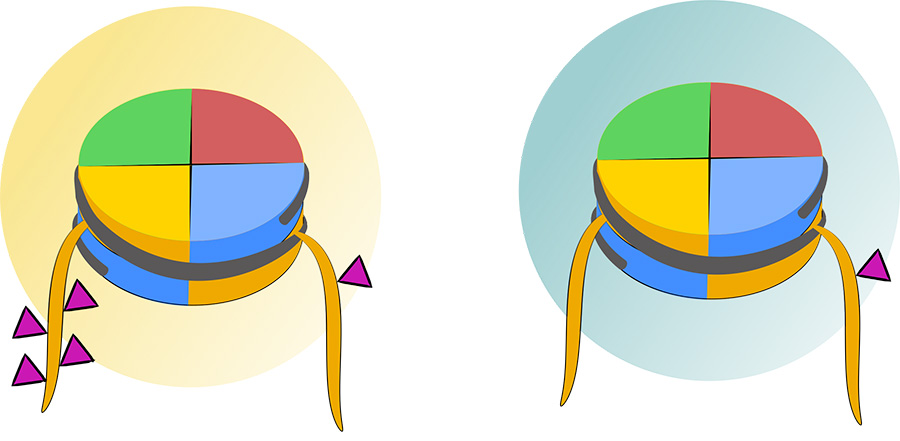
The team knew an enzyme found in bacteria called sortase would cut proteins at a specific protein sequence, and that a similar sequence was present on the tail of histone H3. They selectively engineered the enzyme to target histone H3, generating an intact tail fragment that could be analyzed with mass spectrometry.
They used this new enzyme to isolate H3 tails from human embryonic kidney cells treated with either an HDAC1-specific inhibitor or the CoREST inhibitor corin. Around 200 distinct histone-PTM proteoforms were identified, some of which were unique to the treatment conditions. Cole’s lab is now examining which enzymes are responsible for these modifications, with future research aimed at understanding how the PTMs influence gene expression. Understanding these molecular mechanisms may help identify new pathways to target for disease treatment.
The team also used the sortase enzyme to make what Cole called “designer nucleosomes.” Nucleosomes are comprised of DNA wrapped around a core of eight histone proteins, and the histone tails extend beyond the core structure. The sortase enzyme allows Cole to selectively modify the tails and understand how modifications influence each other.
They found that certain modification patterns can enhance or inhibit co-modifying enzymes, depending on the modification site and enzymes involved. These effects occurred either within the same histone tail or between different tails.
“We think this could have biological significance in how various (enzymes make DNA) more accessible to things like transcriptional machinery,” Cole said.
The team is still investigating the mechanisms of PTM crosstalk, which could lead to new insights into gene regulation and potential therapies for diseases like cancer.
If you missed it, you can watch Cole’s full Breakthroughs webinar here.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.