Flipping the switch
Eukaryotic cells must balance multiple factors before they can grow. The mechanistic target of rapamycin complex 1, or mTORC1, pathway senses and responds to various environmental cues, including nutrient availability, to control protein synthesis and cell growth.
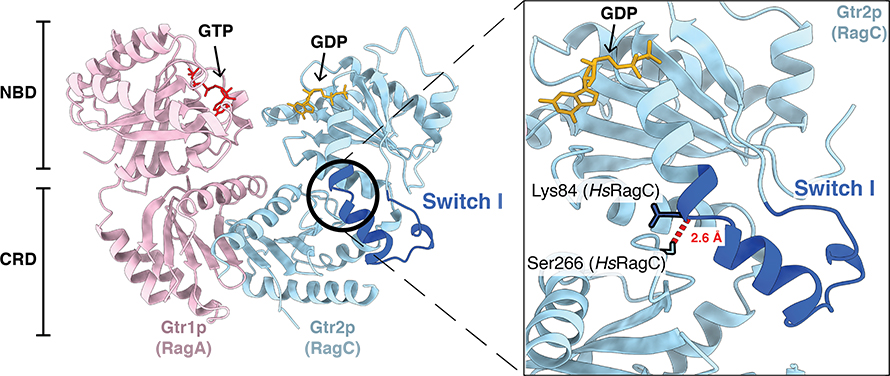
The RagA–RagC GTPase heterodimer is required to activate mTORC1. Depending on amino acid availability, the RagA and RagC subunits will be oppositely loaded with either guanosine triphosphate or guanosine diphosphate, known as GTP and GDP. This variable nucleotide binding induces conformational changes in the heterodimer structure that promote the interaction or release of mTORC1.
When concentrations of amino acids in the cell are high, RagA is bound to GTP, and RagC is bound to GDP — this structural conformation facilitates mTORC1 interaction and activation of protein synthesis at the lysosomal surface. The reverse GTP/GDP loading leads to inhibition of mTORC1 and release from the lysosomal surface into the cytosol.
Until recently, researchers did not understand clearly how RagA and RagC maintain their oppositely nucleotide-loaded state to regulate mTORC1. In an article in the Journal of Biological Chemistry, a team from the University of Massachusetts Chan Medical School describes the molecular mechanism behind this regulation and the consequences of disrupting this balance.
Kuang Shen, an assistant professor at UMass and senior author on the article, described the RagA–RagC complex as a complicated light switch that controls mTORC1 — only when one switch is up and one switch is down will the mTORC1 lightbulb turn on.
“We were curious about the mechanism of regulation, because for us it’s like a black box and we didn’t understand how it worked,” Shen said.
To peer into the black box, graduate student Shawn Egri used enzyme kinetics, structural biology and chemistry tools to probe the mechanism behind the regulation. Egri said he purified between 30 and 40 RagA–RagC mutants with individual amino acid differences and tested their activity one by one.
Eventually, the researchers identified mutants that eliminated normal RagA–RagC activity. They found that one interdomain hydrogen bond is all it takes to keep the switch in one position or the other and prevent it from spontaneously flipping between the two states.
“It was really amazing that just one hydrogen bond has this profound of an impact,” Shen said.
They next introduced mutants in which the interdomain hydrogen bond was broken (meaning the Rag GTPase cannot maintain its oppositely nucleotide-loaded state) into cells. They observed that with the interdomain hydrogen bond abolished, the GTPase subunits could not maintain their original nucleotide-loaded states in cells, leading to distorted responses to amino acid availability and altered mTORC1 interactions.
“It was surprising to us that our results (with purified proteins) translated so strongly in cells,” Egri said. “The phenotype was very clear even in wild-type cell background.”
In the future, Egri and Shen want to lock the complex in the on and off positions to see how that affects interactions with upstream regulators and downstream effectors of the mTORC1 pathway. This could help piece together a better idea of the signals and pathways that regulate the essential processes of protein synthesis and cell growth.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

ASBMB announces 2026 JBC/Tabor awardees
The seven awardees are first authors of outstanding papers published in 2025 in the Journal of Biological Chemistry.

Missing lipid shrinks heart and lowers exercise capacity
Researchers uncovered the essential role of PLAAT1 in maintaining heart cardiolipin, mitochondrial function and energy metabolism, linking this enzyme to exercise capacity and potential cardiovascular disease pathways.

Decoding how bacteria flip host’s molecular switches
Kim Orth will receive the Earl and Thressa Stadtman Distinguished Scientists Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Defining JNKs: Targets for drug discovery
Roger Davis will receive the Bert and Natalie Vallee Award in Biomedical Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.