MicroID2: Streamlined for better biotinylation
The study of protein interactions is a big field in molecular biology. Proteins network with one another for both intercellular and extracellular signaling pathways, many of which require the relevant proteins to be close together.
Researchers often use proximity-dependent biotinylation to characterize these interactions. In this process, an enzyme that labels proteins with biotin is attached to a protein of interest using CRISPR-based or plasmid-based expression strategies. When biotin is added to the experimental system, any protein in close proximity to the protein of interest is tagged and researchers then can purify and identify it.
Proximity-dependent biotinylation recently was used to discover sites where mitochondria contact the endoplasmic reticulum. The outer mitochondrial membrane is difficult to map by purification; the process quickly destroys the membrane, and membrane enrichment techniques are not perfect. By using biotinylation, researchers in the Ting lab at Stanford University accurately captured and identified proteins on the outer mitochondrial and endoplasmic reticulum membranes of living human cells.
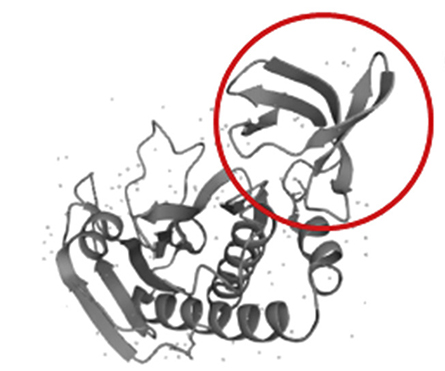
James Londino’s lab at the Ohio State University Wexner Medical Center in Columbus, Ohio, has modified the structure of a state-of-the-art proximity-dependent biotinylation protein, BioID2,that is derived from Aquifex aeolicus bacteria. Benjamin Johnson, a graduate student and first author of a recent paper in the journal Molecular & Cellular Proteomics,explained that by removing the C-terminus of BioID2, the Londino lab created MicroID2, a smaller biotin ligase with fewer nonspecific labeling events than its predecessors.
“We asked, ‘Do we need this entire construct in order to facilitate labeling?’” Londino said.
The size of the ligase matters because larger constructs can cause mislocalization of the protein and disruption of endogenous signaling. This means the host cell will not perform its signaling pathways and other reactions as it would without the insertion of this new protein, skewing the validity of any results. A too-large construct may appear to associate with proteins in artificial ways.
The biotin ligase has a minimum size limit, however. Truncation of the BioID2 C-terminus beyond 63 amino acids completely inactivated the biotinylation activity. Deletion of 10 amino acids from the N-terminus also inactivated the enzyme.
The researchers performed other mutations to develop MicroID2, including substitutions at the active site, that increased the labeling efficiency. Additional mutations reduced nonspecific labeling, allowing more accurate analysis of protein–protein interactions.. Additional mutations reduced nonspecific labeling, allowing more accurate analysis of protein–protein interactions.
“When we’re overexpressing these constructs, they’re going to be continuously labeling,” Londino said. “Even when we completely deplete biotin from the media, there’s still going to be a certain amount of biotin labeling that occurs. By reducing the ability of these constructs to scavenge biotin … we were able to reduce the overall amount of background labeling.”
Johnson described MicroID2 as the smallest biotin ligase yet developed that maintains a high level of activity, but the lab is not done optimizing the construct. Next steps include further exploration of stable integration of the ligase and optimization of its stability.
The Londino lab plans to use these constructs to examine how ubiquitin ligases target specific proteins for degradation. As a lab studying ubiquitin biology, “we are pretty well positioned” to use this tool to examine protein stability, Johnson said.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

