A surprising modification lowers the lipid binding affinity of a membrane trafficking protein
Proteins undergo nonenzymatic modification by reacting with various molecules. For example, cysteine residues react with the alkylating reagent iodoacetamide, and amino groups react with certain carbonyl compounds including succinimidyl esters.
Nonspecific protein modifications in human tissues also are associated with a number of diseases; for example, most of the long-term consequences of diabetes arise from a damaging series of reactions involving blood glucose (an aldehyde) forming covalent bonds to amino groups on various proteins.
Physicians calculate average blood glucose concentrations in diabetic patients by measuring the levels of the glycated protein hemoglobin A1C, a modified form of the red blood cell protein with glucose attached to the N-terminal amino group of the beta-chain. Because such nonenzymatic reactions are typically irreversible, they result in damaged proteins that can lose function and must be removed via proteolytic degradation, a cell’s normal process of breaking down proteins and recycling amino acids.
In a lab, when reactions occur on foreign proteins during bacterial expression, researchers need to design a purification strategy that removes the modified protein. Many modifications do not show up in sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis and thus are easily overlooked.
In a recent study, we identified an endogenous protein modification occurring on a cluster of lysine residues that are central to lipid binding properties in the vesicle trafficking protein granuphilin, or Slp-4. This protein binds reversibly to plasma membranes via a conserved region called a C2 domain, which binds membranes independently of calcium via a large positively charged surface that interacts with negatively charged lipids. At the center of this surface is a cluster of lysines that has a high affinity for the plasma membrane lipid phosphatidylinositol-(4,5)-bisphosphate, or PIP2. We also found that two of these conserved lysines are susceptible to modification by the endogenous bacterial compound phosphogluconolactone, an intermediate in the pentose phosphate pathway. Not surprisingly, the modified protein binds much less strongly to PIP2 lipids than the unmodified protein.
Researchers previously have reported phosphogluconoylation of bacterially expressed proteins but only as a modification on an N-terminal His-tag, never on an internal lysine sidechain. The modified protein manifested as an early-eluting peak during cation-exchange purification, and we identified the site of modification using mass spectrometry following trypsin digestion.
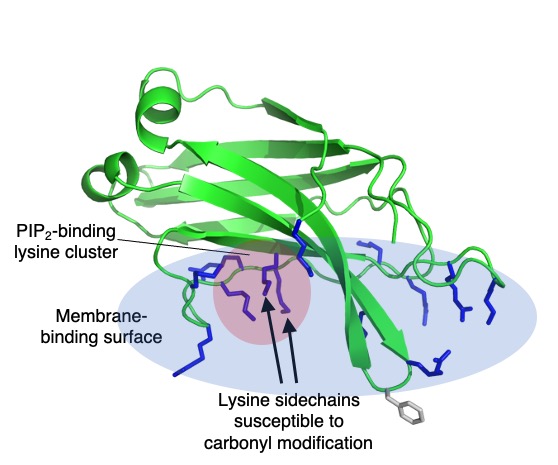
lysine cluster (red circle) embedded in a large electropositive surface (blue oval) containing many basic
residues (blue sticks). A single exposed hydrophobic residue (gray sticks) also contributes to membrane affinity.
Researchers also already knew that certain PIP2-binding C2 domains must be purified via cation exchange in order to obtain reproducible results. Our results suggest that the purpose of this step is not only to remove nucleic acid contaminants but also to separate out this endogenous bacterial modification, which can be a significant percentage of the total protein.
Why is this observation important to lipid biochemistry? Some of the most reactive carbonyl compounds in mammalian cells are aldehydes derived from polyunsaturated fatty acids, or PUFAs. These compounds are downstream products that arise from reaction of PUFAs with reactive oxygen species such as peroxide and superoxide, which become more abundant during oxidative stress and inflammation. PUFAs are especially abundant as acyl chains in phosphoinositide lipids such as PIP2.
Therefore, our observation of reactivity in this PIP2-binding lysine cluster raises several questions: How reactive is this lysine cluster toward endogenous lipid aldehydes in mammalian cells? Does lysine modification affect membrane trafficking? Are there other proteins with lysine clusters that possess similar reactivity?
A serendipitous observation of protein modification during bacterial expression has opened the door to questions at the heart of protein chemistry and membrane biology.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.



