Ocean virus hijacks carbon-storing bacteria
Beneath the ocean’s surface, a virus is hijacking the metabolism of the most abundant organism on Earth. That may be of interest to those of us above who breathe.
Rice University scientists analyzed the role of ferredoxin proteins produced when phages alter the ability of Prochlorococcus marinus to store carbon and counter the greenhouse gas effect arising from fossil fuel consumption.
P. marinus is a photosynthetic cyanobacteria that resides primarily in the tropics and subtropics, where an estimated 10-to-the-27 (an octillion) of them use sunlight to produce oxygen and collectively store four gigatons of carbon annually. Some of this carbon provides critical feedstocks for other marine organisms.
But phages are not their friends. The virus strengthens itself by stealing energy the bacteria produces from light, reprogramming its victim’s genome to alter how it transfers electrons.
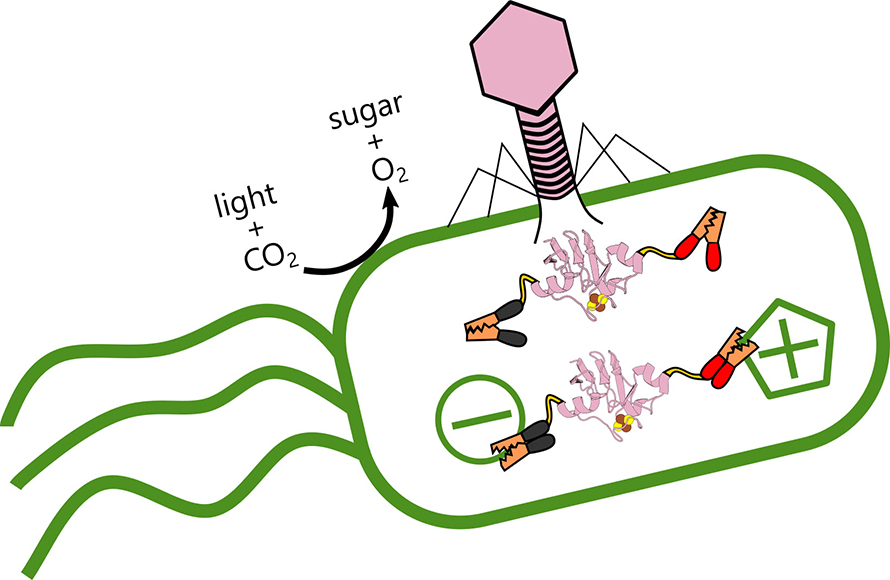
P. marinus and its carbon-storing mechanism are sensitive to temperature, so it bears watching as climate change warms the oceans and extends its range, said Ian Campbell, a Rice postdoctoral researcher and lead author of the study in the Journal of Biological Chemistry.
“The growth in the range of this organism in the oceans could increase the total carbon stored by these microbes,” he said. “Alternatively, the viruses that infect these bacteria could alter carbon fixation and potentially prevent gigatons of carbon from being taken out of the air annually, according to one recent projection.”
Campbell said the goal of the study was to explore the variety of ways viruses interact with their hosts. In the process, the researchers discovered the phage wrests control of electron flow in the host itself, rewiring the bacteria’s metabolism. “When the virus infects, it shuts down production of the bacterial proteins and replaces it with its own variants,” he said. “I compare it to putting a different operating system in a computer.”
The researchers used synthetic biology techniques to mix and match phage and cyanobacterial proteins to study how they interact. A part of the study led by Rice biochemist George Phillips also determined for the first time the structure of a key cyanophage ferredoxin protein.

“A phage would usually go into a cell and kill everything,” said Rice synthetic biologist Jonathan Silberg, the study’s lead scientist and director of the university’s Systems, Synthetic and Physical Biology program.
“But Ian’s results suggest these phages are establishing a complex control mechanism,” he said. “I wouldn’t say they’ve zombified their hosts, because they allow the cells to continue doing some of their own housekeeping. But they’re also plugging in their own ferredoxins, like power cables, to fine tune the electron flow.”
Instead of working directly with cyanophages and P. marinus, Campbell and his team used synthetic biology tools to reprogram much larger, better-understood Escherichia coli bacteria to express genes that mimicked interactions between the two.
“Taking a phage and a cyanobacteria from the ocean and trying to study the biology, especially electron flow, would be really hard to do through classical biochemistry,” Silberg said. “Ian literally took partners from both the phage and the host, put them together by encoding their DNA in another cellular system, and was able to quickly develop some interesting results.
“It’s an interesting application of synthetic biology to understand complex things that would otherwise be arduous to measure,” he said.
The researchers suspect the protein they modeled in E. coli, the Prochlorococcus P-SSM2 phage ferredoxin, is nothing new. “People knew phages encode different things that do electron transfer, but they didn’t know how to connect the wires between the phage and the host,” Silberg said. “They also didn’t know a lot about the phage’s evolution. The structure makes it clear this phage can be traced to specific ancestral proteins involved in photosynthesis.”
Co-authors of the paper are graduate student Jose Luis Olmos Jr., research technician Weijun Xu, postdoctoral researchers Dimithree Kahanda and Joshua Atkinson, undergraduate alumnus Othneil Noble Sparks, research scientist Mitchell Miller, and George Bennett, the E. Dell Butcher Professor of BioSciences and a professor of chemical and biomolecular engineering. Phillips is a professor of biosciences. Silberg is the Stewart Memorial Professor of Biochemistry and a professor of biosciences, bioengineering, and chemical and biomolecular engineering.
The Department of Energy, NASA, the National Science Foundation, the Moore Foundation, the National Cancer Institute and the National Institute of General Medical Sciences supported the research.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

