Untangling the genetics that underlie our facial features
When Eric Mueller, who was adopted, first saw a photograph of his birth mother, he was overcome by how alike their faces were. It was, he wrote, “the first time I ever saw someone who looked like me.” The experience led Mueller, a photographer in Minneapolis, into a three-year project to photograph hundreds of sets of related people, culminating in the book Family Resemblance.
Such resemblances are commonplace, of course — and they point to a strong underlying genetic influence on the face. But the closer scientists look into the genetics of facial features, the more complex the picture gets. Hundreds, if not thousands, of genes affect the shape of the face, in mostly subtle ways that make it nearly impossible to predict a person’s face from examining the impact of each gene in turn.
As researchers learn more, some are starting to conclude that they need to look elsewhere to develop an understanding of faces. “Maybe we’re chasing the wrong thing when we’re trying to create gene-level explanations,” says Benedikt Hallgrímsson, a developmental geneticist and evolutionary anthropologist at the University of Calgary, Canada.
Instead, Hallgrímsson and others think they may be able to group genes into teams that work together as the face forms. Understanding how these teams work, and the developmental processes they affect, should be much more manageable than trying to sort out the effects of hundreds of individual genes. If they’re right, faces may turn out to be less complicated than we think.
Plotting the facial landscape
When geneticists first set out to understand faces, they started with the low-hanging fruit: identifying the genes responsible for facial abnormalities. In the 1990s, for example, they learned that a mutation in one gene causes Crouzon syndrome — characterized by wide-set, often bulging eyes and an underdeveloped upper jaw — while a mutation in a different gene leads to the down-slanting eyes, small lower jaw and cleft palate of Treacher Collins syndrome. It was a start, but such extreme cases said little about why normal faces vary as much as they do.
Then, beginning about a decade ago, geneticists began to take a different approach. First, they quantified thousands of normal faces by identifying landmarks on each person’s face — tip of chin, corners of lips, tip of nose, outside corner of each eye, and so forth — and measuring the distances between them. Then they screened the genomes of those individuals, to see whether any genetic variants corresponded with particular facial measurements, an analysis known as a genome-wide association study, or GWAS (pronounced jee-wass).
Some 25 GWASs of facial shape have been published to date, with over 300 genes identified in total. “Every single region is explained by multiple genes,” says Seth Weinberg, a craniofacial geneticist at the University of Pittsburgh. “There’s some genes pushing outward and others pushing inward. It’s the total balance that ends up becoming you, and what you look like.”
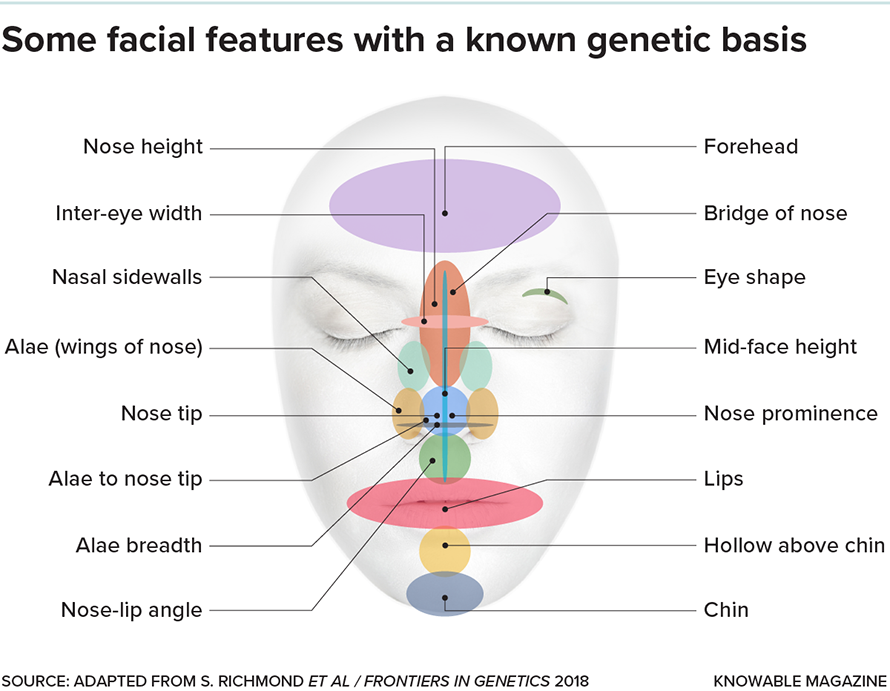
Not only are there a slew of genes involved in each particular facial region, the variants uncovered thus far don’t account well for the specifics of each face. In a survey of the genetics of faces in the 2022 Annual Review of Genomics and Human Genetics, Weinberg and his colleagues gathered GWAS results on the faces of 4,680 people of European ancestry. Known genetic variants explained only about 14 percent of the differences in faces. An individual’s age accounted for 7 percent, sex for 12 percent, and body mass index for about 19 percent of variation, leaving a whopping 48 percent completely unexplained.
Clearly, something important in determining the shape of faces isn’t captured by GWASs. Of course, some portion of that missing variation must be explained by environment — in fact, researchers have noted that certain parts of the face, including the cheeks, lower jaw and mouth, do seem more susceptible to environmental influences such as diet, aging and climate. But another clue to that missing factor, many researchers agree, lies within the unique genetics of individual families.

Variants great and small
If faces were simply the sum of hundreds of tiny genetic effects, as the GWAS results imply, then every child’s face should be a perfect blend, halfway between its two parents, Hallgrímsson says, for the same reason that flipping a coin 300 times will almost always yield roughly 150 heads. Yet you only have to look at certain families to see that’s not the case. “My son has his grandmother’s nose,” says Hallgrímsson. “That must mean there are genetic variants that have large effects within families.”
But if some face genes do have large effects that are visible within the families that carry them, why don’t they show up in a GWAS? Perhaps the variants are too rare in the general population. “Facial shape is really a combination of common and rare variation,” says Peter Claes, an imaging geneticist at KU Leuven in Belgium. As a possible example, he points to French actor Gérard Depardieu’s distinctive nose. “You don’t know the genetics yet, but you feel this is a rare variant,” he says.
A few other distinctive facial features that run in families, such as dimples, cleft chins and unibrows, could also be candidates for such rare, high-impact variants, says Stephen Richmond, an orthodontic researcher at the University of Cardiff, Wales, who studies facial genetics. To look for such rare variants, though, researchers will need to move beyond GWASs to explore large datasets of whole-genome sequences — a task that will have to wait until such sequences, linked to facial measurements, become much more abundant, says Claes.

Another possibility is that the same gene variants that have small effects most of the time could have larger effects within certain families. Hallgrímsson has seen this in mice: He and his colleagues, notably Christopher Percival, now at Stony Brook University, introduced mutations that affect craniofacial shape into three inbred lineages of mice. They discovered that the three lineages ended up with quite different facial shapes. “The same mutation in a different strain of mice can have a different effect, sometimes even the opposite effect,” says Hallgrímsson.
If something similar happens in people, it’s possible that within a particular family — as with a particular strain of mice — the family’s unique genetic background may make certain face shape variants more powerful. But proving that this happens in people, without the aid of inbred strains, is likely to be difficult, Hallgrímsson says.
A better approach, Hallgrímsson thinks, might be to look at the developmental processes that underlie how faces are formed. Developmental processes involve teams of genes that work together — often to regulate the activity of still other genes — to control how specific organs and tissues form during embryonic development. To identify processes linked to face shape, Hallgrímsson and his team first used fancy statistics to find genes that affect craniofacial variation in over 1,100 mice. Then they turned to genetics databases to identify the developmental processes that each gene was a part of. The analysis flagged three processes as especially important: cartilage development, brain growth and bone formation. It’s possible, Hallgrímsson speculates, that individual differences in the rate and timing of these three processes (and likely some others) might be a big part of the explanation for why one person’s face differs from another’s.
Intriguingly, it appears that some of these teams of genes may have “captains” that direct the activity of other team members. Researchers trying to understand facial variation might thus be able to focus on the action of these captain genes rather than hundreds of individual genetic players. Support for this notion comes from an intriguing new study by Sahin Naqvi, a geneticist at Stanford University, and his colleagues.
Naqvi began with a paradox. He knew that most developmental processes are so finely tuned that even modest changes in the activity of the genes regulating them can cause severe developmental problems. But he also knew that small differences in those very same genes are likely the reason his own face looks different from his neighbor’s. How, Naqvi wondered, could both of these ideas be true?
To try to reconcile these two contradictory notions, Naqvi and his colleagues decided to focus on one regulatory gene, SOX9, which controls the activity of many other genes involved in the development of cartilage and other tissues. If a person has only one working copy of SOX9, the result is a craniofacial disorder called Pierre Robin sequence, characterized by an underdeveloped lower jaw and numerous other problems.
Naqvi’s team set out to reduce SOX9 activity little by little and measure what effect that had on the genes it regulates. To do so, they genetically engineered human embryonic cells so that they could dial down SOX9’s regulatory activity at will. Then the researchers measured the effect of six different SOX9 levels on the activity of the other genes. Would the genes under SOX9’s control maintain their activity despite small changes in SOX9, thus keeping development stable, or would their activity decline in proportion to changes in SOX9?
The genes fell into two classes, the team found. Most of them didn’t change their activity unless SOX9 levels fell to 20 percent or less of normal. That is, they seemed to be buffered against even relatively large changes in SOX9. This buffering — possibly the result of other regulatory genes compensating for reductions in SOX9 — would help keep development finely tuned.
But a small subset of the genes turned out to be sensitive to even small changes in SOX9, dialing their own activity up or down in lockstep with it. And those genes, the scientists found, tended to affect jaw size and other facial features altered in Pierre Robin sequence. In fact, these unbuffered genes seem to determine how much, or how little, a regular face resembles a Pierre Robin one. At one end of the range lie the underdeveloped jaw and other structural changes of Pierre Robin sequence. And at the other end? “You can think of the anti-Pierre Robin as an overdeveloped jaw, elongated with a prominent chin — kind of like me, actually,” says Naqvi.
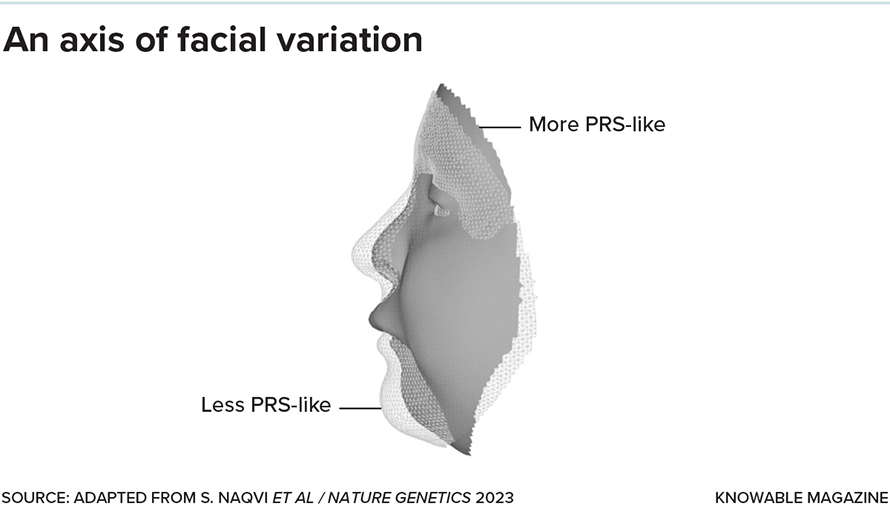
In essence, SOX9 captains a team of genes that define one direction, or axis, in which faces can vary: from more to less Pierre-Robin-like. Naqvi is now looking to see whether other teams of genes, each captained by a different regulatory gene, define additional axes of variation. He suspects, for example, that genes sensitive to small changes in a gene called PAX3 might define an axis relating to the shape of the nose and forehead, while those sensitive to another called TWIST1 — which, when mutated, leads to premature fusing of the skull bones — could define an axis relating to how elongated the skull and forehead are.
Other evidence hints that Naqvi might be on the right track in thinking that faces vary along predefined axes. For example, geneticist Hanne Hoskens, Claes’s former student and now a postdoc in Hallgrímsson’s lab, sorted people’s faces according to how closely they resembled the prominent forehead, flattened nose and other features characteristic of achondroplasia, the most common form of dwarfism. (Think of the actor Peter Dinklage, for example.) Those at the more dwarflike end of the range tended to have different variants of genes related to cartilage development than those with less dwarflike faces, she found.
If similar patterns occur for other developmental pathways, this may set guardrails that restrict the way faces develop. That could help geneticists cut through the complexities to extract broader principles underlying facial shape. “There is a limited set of directions along which faces can vary,” says Hallgrímsson. “There are enough directions that there is a tremendous amount of variation, but it’s a small subset of the geometric possibilities we see. And it’s because these axes are determined by developmental processes, and there are relatively few developmental processes.”
Until more results are in, it’s too early to say whether this new approach really holds an important key to explaining why one person’s face looks different from another’s — and the shock of recognition Eric Mueller experienced when he saw his mother’s picture for the first time. But if Hallgrímsson, Naqvi and their colleagues are on the right track, focusing on developmental pathways may offer a way to part the thicket of hundreds of genes that for so long has obscured our understanding of faces.
Practical applications

If researchers find a way to understand more about the genetic underpinnings that determine the shape of our faces, this could lead to several practical payoffs.
Orthodontists designing a treatment plan for a patient, for example, would like to know how much a young person’s jaw is likely to grow in the years ahead, and whether certain treatments are likely to work. If the patient’s genes could hint at the answer, says Stephen Richmond, an orthodontic researcher at the University of Cardiff in Wales, that would make decisions easier.
Another promising avenue is the use of face shape as a tool in diagnosing other medical conditions. After all, notes Peter Claes of KU Leuven in Belgium, about 40 percent of clinical syndromes — genetic abnormalities that cause multiple defects — involve the face, which suggests that genes that affect the face have many other effects throughout the body. “Based on this,” he says, “I strongly believe the face could be used more in the future as a screening tool.”
For instance, Loeys-Dietz syndrome, a connective tissue disorder that can lead to fatal aortic aneurism, is often hard to diagnose ahead of time, but the syndrome does cause some distinctive facial features that might provide a clue. “If you can do that from the face, with an automated tool that you can put in a family physician’s office, that would be a tremendous benefit,” says Benedikt Hallgrímsson of the University of Calgary, Canada.
— Bob Holmes
This article originally appeared in Knowable Magazine, an independent journalistic endeavor from Annual Reviews.

Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

