MCP: Deciphering the RAF inhibitor paradox
The mitogen-activated protein kinase signaling pathway, called MAPK for short, is a crucial pathway for cellular growth and division and is activated aberrantly in roughly 30 percent of cancers, which is why scientists have sought for drugs that inhibit it. In a study published in the journal Molecular & Cellular Proteomics, researchers at Université de Montréal aimed to delineate the changes that underlie a paradoxical effect seen in cancers that have mutations in a central member of the MAPK pathway. The lead investigator, Pierre Thibault, says, “This research will bring us closer to a real understanding of the biological response of cells to environmental cues and to therapeutic agents.”
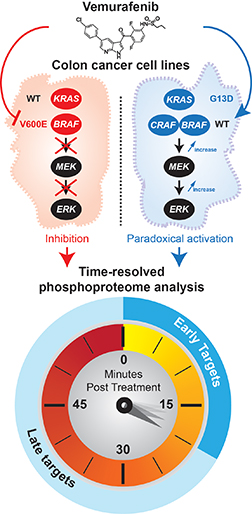 In cancer cells with RAS mutations, vemurafenib activates the MAPK pathway. The research team treated cells with vemurafenib and then subjected them to phosphoproteome analysis. They took snapshots every five minutes for an hour.Image courtesy of Pierre Thibault
In cancer cells with RAS mutations, vemurafenib activates the MAPK pathway. The research team treated cells with vemurafenib and then subjected them to phosphoproteome analysis. They took snapshots every five minutes for an hour.Image courtesy of Pierre Thibault
One class of drugs, a group of inhibitors against the kinase B-RAF, has been effective in stemming the aberrant activation of the MAPK pathway. B-RAF frequently is mutated in cancers; in fact, the V600E mutation of B-RAF is highly prevalent in melanomas and thyroid cancers. Vemurafenib, a drug approved by the U.S. Food and Drug Administration, inhibits this mutant B-RAF. It is especially efficacious in treating cancers with the V600E mutation.
But there’s a paradox. In cancer cells with mutations in an upstream activator of B-RAF called RAS, vemurafenib and inhibitors like it provoke the opposite response — they activate the MAPK pathway instead. This causes drug resistance in tumors and limits the efficacy of the inhibitors. Thibault characterizes this paradoxical pathway activation as “a textbook example of unpredicted events caused by pharmacological agents.”
In collaboration with Marc Therrien at Université de Montréal, Thibault and his team used high-resolution mass spectrometry to profile the temporal changes in the phosphorylation levels of all proteins in the cell, the phosphoproteome, that occur when cancer cells are treated with vemurafenib. They coupled high-resolution mass spectrometry to a method called stable isotope labeling of amino acids in cell culture, or SILAC, which labels cells with heavy versions of amino acids to quantify accurately the changes in protein phosphorylation between the treated and control cells.
A phosphoproteome profile at a time point provides a snapshot of the signaling picture at that time point. However, this static picture limits the information obtained, which is particularly problematic when trying to understand a highly dynamic event, such as cellular signaling activation. So the investigators painstakingly took snapshots every five minutes up to one hour after inhibitor treatment. By doing so, the researchers managed to understand how phosphorylation of proteins changes over time.
The investigators identified 37,910 phosphorylation sites. By comparing the kinetic profiles of two cell lines that harbor either an RAS or a B-RAF mutation, the researchers uncovered which proteins were modulated in response to vemurafenib. They identified 660 phosphorylation sites that displayed temporal response in line with the paradoxical effects in the mutated RAS cancer cells.
From the 660 phosphorylation sites, they discovered a number of novel targets of ERK2, the downstream effector of B-RAF. One of these is TEAD3, a transcription factor involved in the Hippo pathway, another important pathway in cancer, suggesting a direct link between the MAPK pathway and the Hippo pathway. Another novel target found was MKL1, a cofactor involved in cancer metastasis. Through a series of biochemical experiments, the investigators discovered a novel mechanism of MKL1 phosphorylation by ERK2.
“This approach will greatly assist researchers who are developing better inhibitors of B-RAF to inhibit oncogenic signaling in colorectal cancer and other cancer types without adverse paradoxical activation,” says Thibault. He also believes that their approach will be of use to answer questions in basic biology, such as the effects of sustained versus transient ERK activation and how these effects influence the fate of the cell. Thibault says, “Our approach will allow us to tackle these fundamental questions in several cellular contexts.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Decoding how bacteria flip host’s molecular switches
Kim Orth will receive the Earl and Thressa Stadtman Distinguished Scientists Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Defining JNKs: Targets for drug discovery
Roger Davis will receive the Bert and Natalie Vallee Award in Biomedical Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Building better tools to decipher the lipidome
Chemical engineer–turned–biophysicist Matthew Mitsche uses curiosity, coding and creativity to tackle lipid biology, uncovering PNPLA3’s role in fatty liver disease and advancing mass spectrometry tools for studying complex lipid systems.

Redefining lipid biology from droplets to ferroptosis
James Olzmann will receive the ASBMB Avanti Award in Lipids at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Women’s health cannot leave rare diseases behind
A physician living with lymphangioleiomyomatosis and a basic scientist explain why patient-driven, trial-ready research is essential to turning momentum into meaningful progress.

Life in four dimensions: When biology outpaces the brain
Nobel laureate Eric Betzig will discuss his research on information transfer in biology from proteins to organisms at the 2026 ASBMB Annual Meeting.

