JBC: Simulations might speed drug target discovery
Researchers at the California Institute of Technology have developed an approach to overcome a major stumbling block in testing new drug targets. The work was reported in in the Journal of Biological Chemistry.
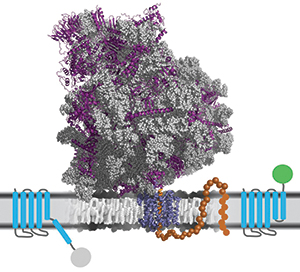 A new method improves protein yield by predicting membrane insertion efficiency,
A new method improves protein yield by predicting membrane insertion efficiency,a key step in the expression of membrane protein. courtesy of Thomas Miller III and William Clemons Jr./caltech
Proteins embedded in cell membranes are potential targets for drugs to treat a number of diseases, from infectious diseases to cancers. Membrane proteins (which include transporters, channels and receptors) are the targets of almost 70 percent of FDA-approved drugs.
However, it is notoriously difficult for researchers to produce membrane proteins in the lab in sufficient quantities to be able to purify them and conduct experiments with potential drugs. Thomas F. Miller III and William M. Clemons Jr. of the department of chemistry and chemical engineering at Caltech wondered whether there was a way to help researchers experiencing this problem.
“Our motivation for this project was really born out of frustration with this general problem, which is that membrane proteins are very hard to produce at scale for experimental purposes,” Clemons said.
To produce proteins of interest, researchers typically insert the gene encoding the protein into a laboratory workhorse cell line, such as Escherichia coli; this process is called heterologous overexpression of a protein. But membrane proteins typically are overexpressed in only very small amounts for reasons that have been poorly understood until now. Individual researchers sometimes spend years trying to modify their proteins of interest in ways that will make them more efficiently expressed in the lab.
“People just hunt around in the dark to hopefully find something that works better so that they can get enough protein to perform their studies,” Miller said. “New tools are needed to rationally enhance that, to do it in a more purposeful way.”
To see whether there were any general principles that could guide attempts to improve membrane protein expression, Clemons and Miller and their graduate students Michiel J.M. Niesen and Stephen S. Marshall focused on a specific step in the process: the point when a cell actually inserts a newly synthesized protein into the membrane.
The efficiency of insertion — that is, the fraction of the time that a protein is inserted into the membrane correctly — depends on the protein’s amino acid sequence. The team developed a computational simulation method to predict how a change in the sequence would affect insertion efficiency.
In the new study, the team tested how this predicted efficiency related to protein expression in the lab. The team systematically produced many variants of a particular protein and used the algorithm to predict each variant’s membrane insertion efficiency. Then the researchers quantified how much protein was produced. As they had hypothesized, improved insertion efficiency correlated with improved protein yield.
Now researchers interested in studying a particular membrane protein can use these simulation tools to predict what changes they should make to their protein sequence in order to produce the membrane protein in the lab. There are caveats: If a particular protein in a particular cell type is subject to inefficiencies at steps in its synthesis other than membrane insertion, then the new method may not help. But the researchers are confident that the method offers a way forward for many membrane protein researchers struggling to express their proteins.
“We believe that the tools we’ve developed here have the potential to really revolutionize membrane protein expression,” Clemons said. “There are still things we have to do to fully realize that, but this paper demonstrates that the potential is there.”
The researchers are teaming up with others to put these tools to work.
“There are many membrane protein targets that are of real importance and real value for pharmaceutical and drug design purposes,” Miller said. “If we can help people by bringing an elusive target within grasp, it would be a big victory.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

.jpg?lang=en-US&width=300&height=300&ext=.jpg)