Flipping lipids to deform membranes
Cell membranes change shape during such functions as membrane trafficking, cell migration and cell division. Rearrangement of cytoskeletons and interactions of various proteins, including membrane-bending proteins, regulate these changes. When the composition and asymmetric distribution of lipids between the two leaflets of the membrane bilayer change, membranes are deformed, a requirement for processes such as immune response and nutrient uptake.
In mammalian cells, the phospholipids phosphatidylserine and phosphatidylethanolamine are abundant in the cytoplasmic leaflet of the bilayer, whereas phosphatidylcholine, or PC, and sphingomyelin are abundant in the exoplasmic leaflet. P4-ATPases, also called flippases, catalyze unidirectional flipping of lipid molecules from the exoplasmic to the cytoplasmic leaflets of the bilayer and are crucial for membrane trafficking, but we don’t know how they are coupled to the process of vesicle formation.
P4-ATPase–mediated phospholipid translocation might cause an imbalance of lipid mass between the leaflets of the bilayer, so our lab wanted to find out whether the flipping enzymes of the P4-ATPase family contribute to changes in the shape of cell membranes. PC is abundant in the outer leaflet, so we hypothesized that greater PC-flipping activity causes an increase in lipid mass in the inner leaflet of the plasma membrane and induces inward membrane curvature.
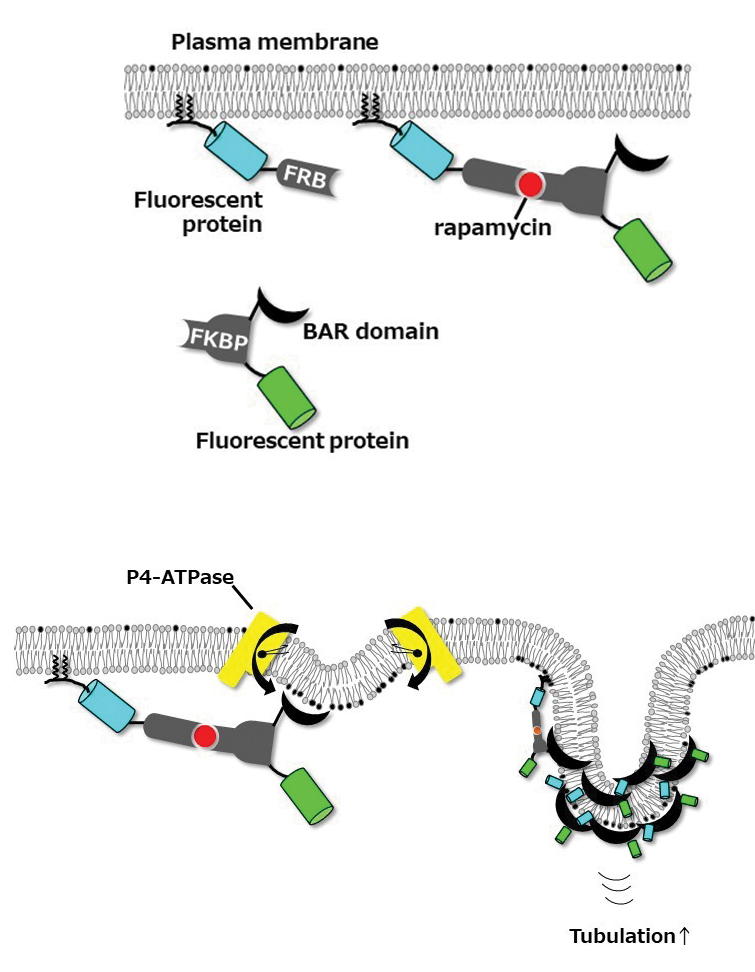
To test this hypothesis, we used BIN/Amphiphysin/Rvs, or BAR, domains, which can sense membrane curvature and induce tubular membrane structures via self-oligomerization. We examined plasma membrane-bending caused by BAR domains with a rapamycin-inducible dimerization system, using FK506 binding proteins, or FKBPs, and FKBP–rapamycin binding domain, or FRB, proteins, that allow for acute recruitment of BAR domains to the plasma membrane. FRB was targeted to the plasma membrane by adding the N-terminal 11 amino acids of Lyn kinase, and FKBP was fused to the BAR domain.
When one specific BAR domain, called N-BAR, is recruited to the plasma membrane, we know that it penetrates the lipid bilayer via its N-terminal amphipathic helix, causing a small inward curve to form. The protein senses this change in curvature, leading to the recruitment of more N-BAR domains, which oligomerize along a part of the membrane and trigger transformation of the membrane into a tube.
Delta-N-BAR (which lacks the N-terminal amphipathic helix of the N-BAR domain) and F-BAR domains did not do this unless ATP10A was expressed exogenously in cells. In 2015, we had published in the Journal of Biological Chemistry our finding that ATP10A flipped PC from the outer to the inner leaflet of the plasma membrane, slightly changing the shape of the plasma membrane. Delta-N-BAR and F-BAR domains sensed the change and bound to the plasma membrane. The proteins then oligomerized along the membrane and transformed that part of the membrane into an inwardly protruding tubule.
Our recent study shows that changes in the transbilayer lipid compositions induced by P4-ATPase can deform biological membranes. Increased inward plasma membrane bending by ATP10A expression enhances endocytosis. The plasma membrane also changes shape during cell migration, cancer cell invasion, cell division, nutrient uptake and entry of pathogens into cells, so lipid-flipping activity may be involved in any or all of these processes.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

ASBMB announces 2026 JBC/Tabor awardees
The seven awardees are first authors of outstanding papers published in 2025 in the Journal of Biological Chemistry.

Missing lipid shrinks heart and lowers exercise capacity
Researchers uncovered the essential role of PLAAT1 in maintaining heart cardiolipin, mitochondrial function and energy metabolism, linking this enzyme to exercise capacity and potential cardiovascular disease pathways.

Decoding how bacteria flip host’s molecular switches
Kim Orth will receive the Earl and Thressa Stadtman Distinguished Scientists Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Defining JNKs: Targets for drug discovery
Roger Davis will receive the Bert and Natalie Vallee Award in Biomedical Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.



