Turning yeast cells into labs for studying drivers of gene regulation
Researchers have developed a more efficient platform for studying proteins that play a key role in regulating gene expression. The approach uses engineered yeast cells to produce enzyme and histone proteins, conduct biochemical assays internally, and then display the results.
“Biomedical and biotech researchers are interested in the mechanisms that allow histones to regulate gene activity,” said Alison Waldman, first author of a paper on the work and a Ph.D. student at North Carolina State University. “But the conventional tools for histone research are unwieldy and slow. We wanted to develop something faster and less expensive — and we did.”
Specifically, the researchers have created a scalable platform that enables efficient characterization of histone modifications without the need for recombinant protein production.
In complex organisms, chromosomes are largely made up of DNA and a group of proteins called histones. These histones are important for packing the DNA into chromosomes properly, but they also play a role in regulating gene expression. In other words, they help determine when and how specific genes are turned on or off.
One of the features that makes histones challenging to study is that they often have chemical modifications that, alone or in combination, alter the role the histone plays in gene expression.

“Histones essentially serve as docking sites for other proteins that influence gene expression, and the chemical modifications we see on histones play a role in determining which proteins have access to a given gene,” said Balaji Rao, co-corresponding author of the paper and a professor of chemical and biomolecular engineering at NC State.
And to make matters more complicated, this is a dynamic process. A histone may have no modifications, it may retain a modification for the entire life of the cell, or modifications may be added and removed repeatedly. There is, in short, a lot going on. And enzymes are the catalysts responsible for all of those changes. Basically, enzymes are the mechanism for attaching or removing histone modifications.
So if you really want to understand what's going on with histones, you have to understand the chemical modifications. But if you want to understand the chemical modifications, you need to understand which enzymes are present and what they are doing.
Conventional methods for understanding how enzymes modify a histone involve using one of two techniques. First, you could use chemical synthesis to create enzymes and histone proteins, then conduct an assay in a test tube to see what happens. Second, you could genetically engineer one bacteria to produce an enzyme and engineer other bacteria to produce histone proteins, then harvest the relevant proteins, purify them, then conduct an assay to see what happens.

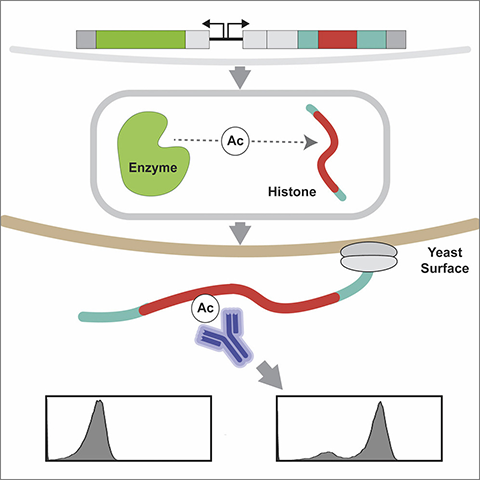
“Our technique uses a genetically modified yeast cell to produce both the enzyme and the histone,” Waldman said. “The chemical modification takes place within the cell, and the resulting modified histone is sent to and displayed on the surface of the cell.”
This is achieved by fusing the histone and enzyme to peptide tags that recruit and retain them in the endoplasmic reticulum, bringing them in close mutual proximity for any enzymatic reaction to occur. The histone is also fused to a yeast surface display protein, AGA2p. This results in the enzyme-modified histone being displayed on the yeast surface, where it can easily be assayed using antibodies against specific histone modifications of interest.
“In other words, the yeast cell makes the relevant proteins, does the assay for you, and then displays the result on top,” said Albert Keung, co-corresponding author of the work and an assistant professor of chemical and biomolecular engineering at NC State.

The modified yeast platform is significantly faster than conventional techniques. For example, examining a single enzyme–histone pairing would take a couple days instead of a week.
“But it’s easier to scale up than existing techniques, so you would save substantially more time if you were looking at a lot of proteins,” Keung said.
“In addition, there are some proteins that can’t be made using chemical synthesis or that can’t be purified,” Rao said. “Our technique doesn’t require chemical synthesis or purification, which means we can look at proteins that were difficult or impossible to assay in the past.”
The researchers demonstrated the utility of the technique by having engineered yeast cells produce two types of histones and a well-studied enzyme called p300, which adds a specific acetyl group modification to histones.
The researchers rapidly profiled the lysine residues on histones H3 and H4 that human p300 acetylates by antibody labeling and flow cytometry analysis of intact live yeast. Because there are widespread issues with the accuracy and specificity of commercial antibodies, the researchers also modified the experimental platform in two ways to rapidly and clearly identify which antibodies were working as expected and as advertised by their vendors. First, the researchers mutated each lysine residue to determine if the antibody labeling was ablated; second, they linked chemically synthesized peptides to the yeast surface through a streptavidin–biotin interaction.
The scalability and efficiency of the platform was further demonstrated by mapping the large binary crosstalk space between acetylated residues on histones H3 and H4, uncovering previously unreported residue interdependencies affecting p300 activity. To do this, each histone H3 and H4 lysine was individually mutated to an arginine, and the effect of each mutation on the acetylation of all other histone lysines was profiled.
“We’ve shown that our technique works,” Waldman said. “The next step is to begin expanding the modifications we’re looking at and scaling up the process.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

