Molecular basis for interaction between an essential protein complex and its regulator
The labs of Lauren Jackson, associate professor of biological sciences and biochemistry, and Todd Graham, Stevenson Professor of Biological Sciences at the College of Arts and Science and professor of cell and developmental biology, recently published a study in the Journal of Cell Biology describing a significant interaction between an essential protein complex used for protein and lipid transport—the COPI complex—and its regulator protein.
The authors include Betty Xie and Cameron Cohen, two graduate students in the Jackson lab, and Swapneeta Date, senior scientist in the Graham lab. Together, the two groups established how COPI interacts with the regulator protein Glo3, a member of the ArfGAP protein family of enzymes. Disruptions of the interaction between COPI and Glo3 lead to deleterious effects in cells.
We sat down with Jackson to find out more about this research.

What issue/problem does your research address?
Our lab studies the cellular “FedEx system” that allows proteins and lipids to move between membrane-bound compartments. This work focused on COPI, which is required in all eukaryotes. Mutations in COPI are associated with multiple human diseases, including cancers and microcephalies, which are conditions in which a patient’s head is abnormally small. Pathogens, including viruses like SARS-CoV-2, also hijack COPI to take over cells.
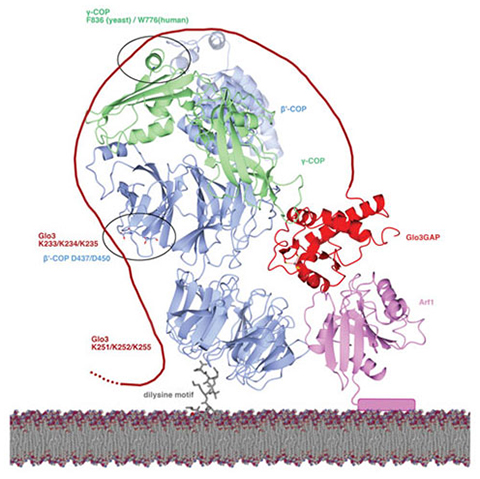
While the field has previously established that molecules in the ArfGAP family affect COPI function, the mechanisms involved in this interaction have not been well understood. We sought to determine the basis for the interaction between COPI subunits and a specific ArfGAP protein, Glo3.
What was unique about your approach to the research?
We undertook a multi-disciplinary approach to address these questions using biochemistry, biophysics, computational artificial intelligence-based modeling, and in vivo approaches including fluorescence imaging in budding yeast. This combination of approaches allows us to use the molecular information generated from biochemical, biophysical, and computational studies to design hypotheses that could be used to test function directly in budding yeast, our model organism.
What were your findings?
We discovered a molecular “code” that explains how COPI recruits ArfGAP proteins. We identified specific amino acid residues on each protein that mediate the interaction between COPI and ArfGAP. We then tested how breaking the interaction affected the movement of transmembrane protein cargo using budding yeast as our model organism. Breaking this interaction specifically disrupted the shape of a key organelle called the Golgi, and it also misdirected specific cargo proteins to the wrong cellular location.
What do you hope will be achieved with the research results in the short and long terms?
Overall, many of these proteins are strongly conserved across species, which suggests our results will be relevant to human biology. This study provides a variety of molecular tools that we can use to dissect these important cellular events that drive essential membrane trafficking—the process of moving proteins and other molecules throughout the cell using membrane-bound vesicles.
What are the benefits of this research?
This research contributes to our understanding of fundamental biological processes in cellular sorting. Obtaining molecular information allows us to probe complex cellular sorting pathways so that we can understand human physiology and what goes wrong in disease states, such as in neurological disorders or cancers, when essential proteins are mutated or lost
Where is this research taking you next?
This work builds on our ongoing collaboration with Todd Graham’s lab. Together, the Jackson and Graham labs use a variety of methods to understand how the COPI/Glo3 interaction drives specific cellular sorting events (Jackson lab) and how the interaction is affected by post-translational modifications and sorting of SNAREs, a family of proteins that play essential roles in vesicle fusion events (Graham lab).
This article was written for Vanderbilt University’s School of Medicine Basic Sciences and was republished with permission. Read the original.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

ASBMB announces 2026 JBC/Tabor awardees
The seven awardees are first authors of outstanding papers published in 2025 in the Journal of Biological Chemistry.

