Unveiling an elusive microbial enzyme
In the 1980s, scientists isolated a novel protein from the common airborne fungus Aspergillus fumigatus. Initial studies described this enzyme’s functions, but the gene encoding it remained unknown for 40 years until it recently was identified by researchers at the University of Tokyo. Graduate student Toma Kashima and collaborators published this work in the Journal of Biological Chemistry, providing an extensive biochemical characterization of this enzyme.
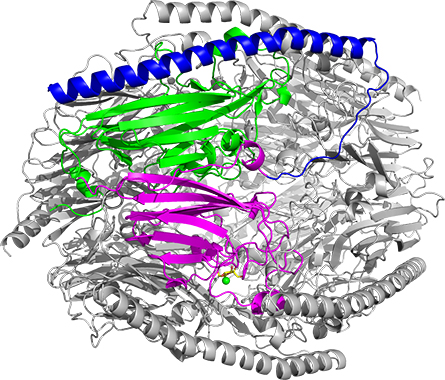
Prebiotics are nondigestible substances that can help shape specific microbial populations within the digestive tract. They are frequently dietary fibers from plant food sources but can include other compounds. Because mammals lack the enzymes required to break down the bonds within prebiotics, we rely on the bacteria that reside in our gut to do this work. Specifically, a group of enzymes in bacteria known as glycoside hydrolases, or GHs, play a significant role in this process. The University of Tokyo researchers became interested in these GH enzymes within the genome of Bifidobacterium dentium, a bacterium that colonizes the human intestine and oral cavity.
“We first noticed a hypothetical gene next to a GH32 protein and predicted this unknown gene must have a similar function,” said Shinya Fushinobu, corresponding author of this paper.
To begin teasing out the function of this region of the bacterial genome, Fushinobu’s team isolated the protein product of this gene and tested what substances it reacts with using thin-layer chromatography, a method that identifies components of a mixture and follows the progression of a reaction.
“The result was very surprising … with a specific shift in one spot that we did not expect to be there,” Fushinobu said.
The team discovered that this protein had two distinct enzymatic activities, acting as both an alpha-D-fructofuranosidase and an alpha-D-arabinofuranosidase. These two activities break down specific bonds in sugars, the former targeting bonds found only in cooked substances, such as sucrose caramel. Previously studied GH proteins break down b-linked molecules (which hold sugars together), but the researchers found that these two activities acted on a-linked sugars, an area that is not well understood.
“This activity was unexpected because a-bonds and b-bonds are totally different, and cleavage of a-D-fructofuranoside bonds by a GH protein is new,” Fushinobu said.
Following these initial findings and 40 years after its initial discovery, the researchers designated the protein isolated from Bifidobacteriumas aFFase1.
Fushinobu’s team advanced their investigation by determining that aFFase1 has a hexameric structure. They proceeded to mutagenize specific sites within the protein that were revealed to be responsible for binding and reacting with substrates. Collectively, the team assembled their data and proposed a mechanism for how aFFase1 converts particular sugars in foods we consume.
Although there are over 160 recognized GH families, Bifidobacterium’s aFFase1 does not resemble any known amino acid sequence. Its unique features have prompted the creation of a new glycoside hydrolase family, GH172. After elucidating aFFase1’s role in breaking down distinct a-linked bonds within sugars, this research team will explore further the metabolic systems in B. dentium that exploit aFFase1.
“We unexpectedly revisited a very old enzyme discovered by Japanese researchers in the 1980s and found very surprising results,” Fushinobu said. “We believe this new gene will be involved in important degradation systems that can be broadly used in commercial products and give us insights for our health and the bacterial communities in the human gut.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

