Review delves into proximity proteomics
In a recent review article in Molecular & Cellular Proteomics, Payman Samavarchi–Tehrani and colleagues in the Gingras lab at Sinai Health Systems and the University of Toronto offer an introduction to proximity-dependent biotinylation, a key first step in proximity proteomics. The authors give researchers who are new to the field information about the natural history of biotinylation enzymes. They also offer insights into the mechanisms of these enzymes and new perspectives on future proximity proteomics experiments.
Traditional proteomics can provide information about the quantitative contents of a cell or tissue, but it sacrifices much information on the spatial organization of proteins within cells. Since protein activity often depends on location and interactions with other proteins, researchers have developed approaches such as proximity proteomics to obtain information about the environs of a protein of interest. Proximity proteomics methods developed in the past 10 years depend on fusing the protein of interest to an enzyme that will label nearby proteins with a chemical tag that then can be purified. After purification, mass spectrometry identifies the tagged proteins.
Most often, the chemical tag is biotin, a cofactor that is key to carboxylase enzyme activity in several metabolic pathways. Two types of enzyme are used for proximity-dependent biotinylation: peroxidases, used for methods such as APEX, and biotin ligases, used for methods such as BioID.
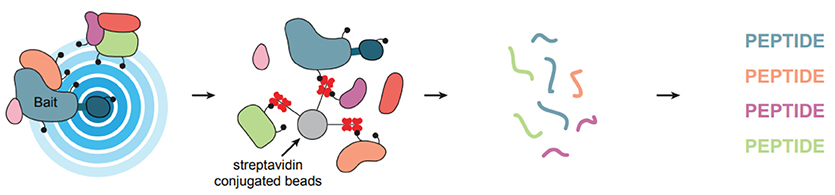
Ordinarily, biotin ligases append biotin to the carboxylases that need it as a cofactor. Biotin ligases found in cells have high specificity for their substrate proteins, but certain mutations reduce that specificity by decreasing the ligase enzyme’s affinity for a reactive intermediate. Such mutants lose their grip on the cofactor and can release a reactive biotin that can bind the next amine group it encounters — often on a nearby protein. When researchers pull down biotin after this reaction occurs, they can determine what proteins were localized in the neighborhood of the biotin ligase and, by extension, the protein it was tethered to.
The second enzyme family, the peroxidases, evolved to convert hydrogen peroxide to water by redox chemistry. In the presence of a biotin–phenol substrate and hydrogen peroxide, they can make a short-lived free radical that reacts with certain amino acid side chains, once again tagging nearby proteins for later identification.
As proximity proteomics has grown in popularity, both types of enzyme have been the targets of extensive engineering and molecular evolution to coax them toward the activity profiles users want. The authors review the available enzymes and discuss experimental design considerations, such as choice of control conditions and how to get rid of what they call “frequent flyer” proteins that often are isolated nonspecifically.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

