From the journals: MCP
Using automated UbiFast to profile ubiquitination. Picturing light, dark and protein degradation. Identifying an enzyme that promotes pancreatic cancer. Read about papers on these topics recently published in the journal Molecular & Cellular Proteomics.
Automated profiling of ubiquitination
Ubiquitylation is a post-translational protein modification that can regulate a variety of cellular functions including protein turnover through the ubiquitin–proteasome system. Dysregulation of ubiquitylation enzymes can lead to activation or deactivation of pathways implicated in diseases, including cancer and neurodegeneration. Some treatments targeting these pathways exist; however, to develop new therapeutics, researchers need a better understanding of how ubiquitin modification influences disease.
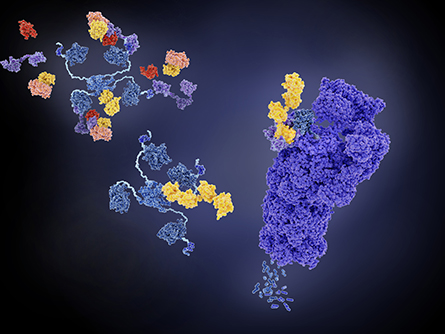
proteosome (right, violet) and can regulate pathways implicated in disease formation.
A team led by Keith Rivera of the Broad Institute recently developed an automated adaptation of UbiFast — a method to identify protein ubiquitylation sites in a high-throughput and multiplexed manner — and this work was published in the journal Molecular & Cellular Proteomics.
With UbiFast, protein samples from cells or tissues are cleaved to peptide fragments, and an antibody is used to enrich for ubiquitin modification sites. Chemical tags are coupled with the antibody enrichment to label samples individually. Labeled peptides from multiple samples can be analyzed in one run using liquid chromatography coupled to mass spectrometry to identify the modified proteins.
In their new method, magnetic beads coupled to the antibody allow sample processing steps, including washing and tagging, to be automated using a 96-well plate format in a robotic magnetic particle processor. This increased sample throughput and also decreased sample processing time and variability across experimental replicates.
To compare the manual and automated UbiFast methods, the group used patient-derived breast cancer xenograft tissue; they found that automation significantly increased depth of coverage of the ubiquitylome.
In addition to providing fast, high-throughput analysis of ubiquitylation, the protocol can be adapted to study other post-translational modifications to cellular processes, such as sumoylation or phosphorylation. Given the role post-translational modifications play in homeostasis and disease, higher-throughput experiments can advance understanding of cellular processes and better identify potential therapeutic targets.
Light, dark and protein degradation
Light is essential for photosynthetic organisms. It provides energy and can regulate cellular processes including protein expression and degradation. Yet researchers lacked a global picture of which proteins are targeted for degradation by light and how they are targeted until a recent study by a team of researchers from China and the U.S. led by Weiyang Chen of the Chinese Academy of Sciences. In their work, published in the journal Molecular & Cellular Proteomics, the researchers studied light-regulated protein degradation events in the photosynthetic organism Synechocystis, a strain of single-cell cyanobacteria that resemble chloroplasts in plants. They used quantitative proteomics to identify proteins with degradation induced by light and also by dark.
The team found that the light-dependent degradation was regulated by the redox status of the intracellular environment. Light-dependent degradation favored a reducing environment that may serve as a more general mechanism underlying light-dependent protein degradation. Their results expand our knowledge of the light-regulated protein quality control mechanism in photosynthetic organisms, which is critical for maintaining intracellular homeostasis during day–night alternation. Photosynthetic organisms are the dominant contributors of oxygen to Earth’s atmosphere and provide much of the food humans consume, so understanding their quality control systems is important.
IDing a cancer-promoting enzyme
Pancreatic ductal adenocarcinoma, or PDAC, is the most common type of pancreatic cancer and the third leading cause of cancer-related death in the U.S. Glycans, sugar-based polymers that coat cells and decorate proteins, are crucial for modulating biological processes and are emerging as important biomarkers of PDAC and other cancers. Recently, Emma Kurz, Shuhui Chen and a team of researchers from New York University analyzed the glycan content in a mouse modified to develop early-stage pancreatic cancer and in human PDAC patient samples to identify glycans involved in cancer formation.
In their study, recently published in the journal Molecular & Cellular Proteomics, the team found that an enzyme called ST6GAL1 that transfers sialic acid to glycans is overexpressed in human PDAC cells. They next tested the role of ST6GAL1 in the mouse model of pancreatic cancer by knocking out ST6GAL1 in the pancreas. With ST6GAL1 deleted, the researchers observed delayed cancer formation and a decrease in the fibrosis that is a hallmark of pancreatic cancer, suggesting ST6GAL1 plays a role in promoting PDAC. This work highlights how systems-based approaches can be used to identify glycans of interest that subsequently can be tested in clinically relevant disease models. It also paves the way for future work to target glycosylation enzymes therapeutically in pancreatic cancer.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

The data that did not fit
Brent Stockwell’s perseverance and work on the small molecule erastin led to the identification of ferroptosis, a regulated form of cell death with implications for cancer, neurodegeneration and infection.

Building a career in nutrition across continents
Driven by past women in science, Kazi Sarjana Safain left Bangladesh and pursued a scientific career in the U.S.

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.