How 'master regulators' of cells make DNA accessible for gene expression
New insight into proteins called “pioneer factors” helps to explain their unusual ability to open up the typically dense genetic material within our cells. This behavior makes the genetic material accessible for proteins involved in important cellular processes, such as DNA replication and repair, gene expression, and the creation of proteins. The study, by a team of researchers from Penn State and the Ohio State University, used a unique combination of structural biology, biophysics, and cell biology to understand how these “master regulators” of the genome interact with nucleosomes — the basic unit of the genome in all eukaryotic cells, which range from yeast to humans.
DNA inside the nucleus of cells is generally wrapped around proteins called histones and packed into dense complexes called nucleosomes. The nucleosomes, which resemble beads on a string of DNA, are further packed together forming chromatin, which then make up chromosomes.
“Nucleosomes are a barrier for a lot of proteins to associate with chromatin, but pioneer factors have a special property that they can invade into the nucleosome and generate open regions that can be accessed by other factors,” said Lu Bai, associate professor of biochemistry and molecular biology and of physics at Penn State, and one of the leaders of the research team. “Because of this, they are sometimes thought of as ‘master regulators’ of genes. In this study, we used a combination of approaches to better understand this how pioneer factors can invade the nucleosome.”
The study appears in the April 20 issue of the journal Molecular Cell.
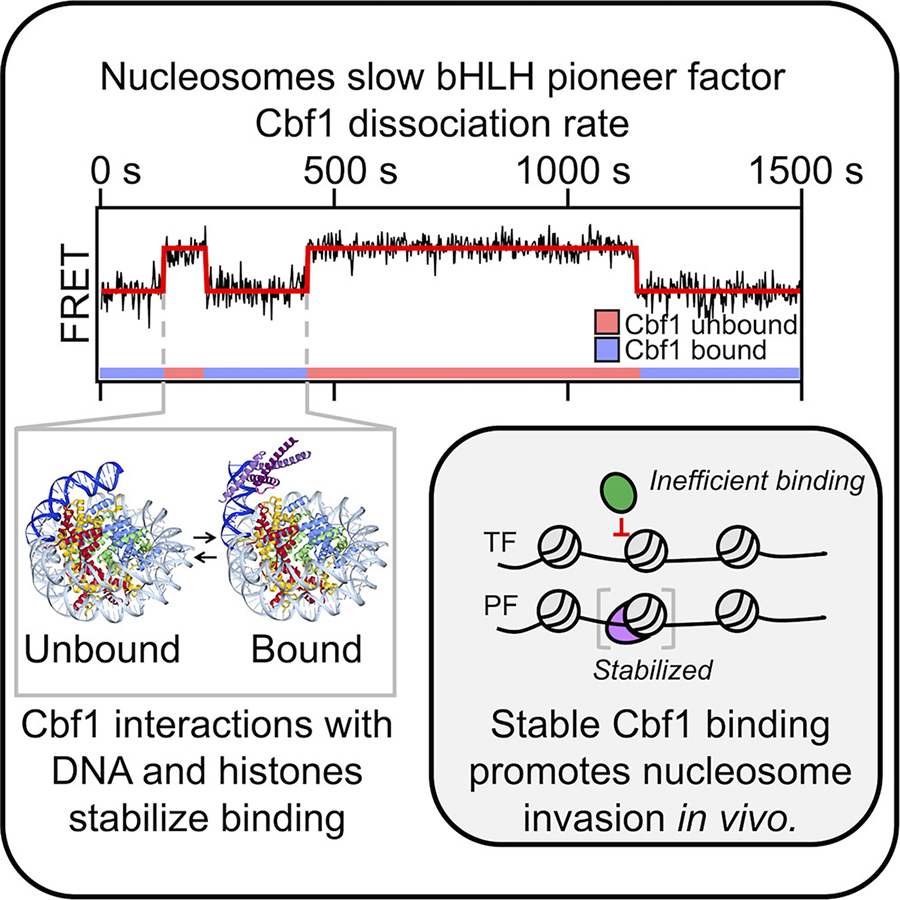
Pioneer factors are a type of transcription factor — proteins critical to the process of transcription, where DNA is copied into the RNA blueprints for the creation of proteins. While many transcription factors can bind to the nucleosome, most fall off very quickly. By contrast, pioneer factors have what is called a “dissociation compensation mechanism” that allows them to remain stably bound to the nucleosome for an extended period of time.
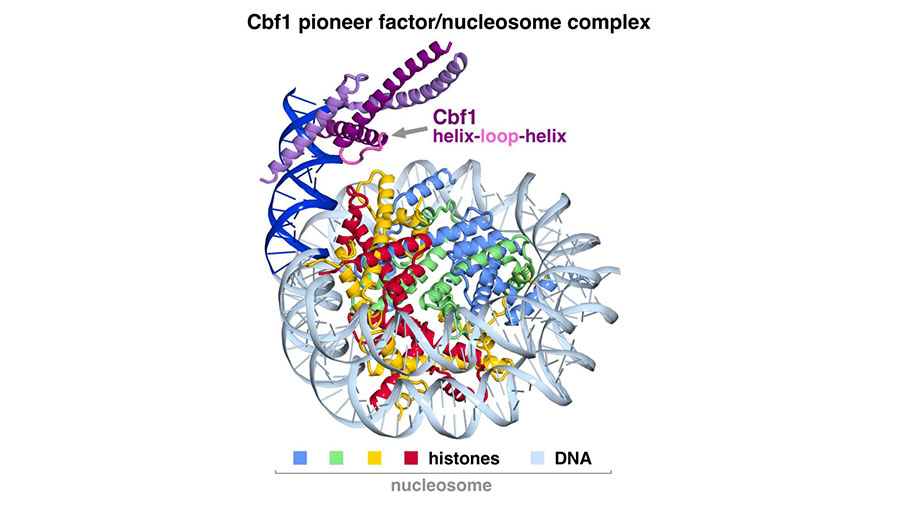
The research team compared a pioneer factor called Cbf1 and a non-pioneer transcription factor called Pho4 in budding yeast. The two proteins have similar overall structures and the ability to recognize the same DNA sequences but they behave differently around the nucleosome. Using a sensitive imaging technique called cryo-electron microscopy, the researchers identified a structure on the pioneer factor that they believe contributes to its ability to invade the nucleosome.
“Based on our cryoelectron microscopy structure, Cbf1 interacts not only with the DNA part of the nucleosome, but also with histones within the nucleosome through its helix-loop-helix region,” said Song Tan, Verne M. Willaman Professor of Molecular Biology at Penn State and one of the leaders of the research team. “We suspect this interaction helps prevent Cbf1 from dissociating as quickly as non-pioneer factors.”
To confirm the role of the helix-loop-helix region, the research team made “chimeras” of each of the proteins, removing a helix-loop-helix from the pioneer factor and adding one to the non-pioneer transcription factor. Then they measure how quickly these chimeras dissociated from the nucleosome.
“Removing the helix-loop-helix from the pioneer factor caused quicker dissociation, which resulted in a reduced dissociation compensation phenomenon and a dramatically less effective pioneer factor,” said Michael Poirier, professor and chair of physics at Ohio State and one of the leaders of the research team. “Amazingly, adding the helix-loop-helix region conferred pioneer factor properties to the non-pioneer factor.”
To further confirm that their results, the researchers explored how these chimeras functioned within live yeast cells. They found that the pioneer factor and non-pioneer chimera modified to behave like a pioneer factor both facilitated invasion and opening up of the nucleosome. These results collectively help explain how pioneer factors like Cbf1 can access the nucleosome and increase accessibility of DNA for other factors.
“This study would not have been possible without combining expertise from our three separate groups,” said Tan “My group focused on structural biology, Michael Poirier’s group at Ohio State focused on single molecule biophysics, and Lu Bai’s group at Penn State focused on in vivo cellular biology. This has been a wonderfully synergistic collaboration and has produced ideas and results that would not have been generated with one or even two of our groups on their own. We are continuing this collaboration to explore interactions of other pioneer factors and the nucleosome.”
In addition to Tan and Bai, the research team in the Center for Eukaryotic Gene Regulation at Penn State includes graduate student Hengye Chen and postdoctoral scholar at the time of the research Priit Eek. In addition to Poirier, the research team at Ohio State includes Benjamin Donavan, Zhiyuan Meng and Caroline Jipa. This work was supported by the U.S. National Institutes of Health, the Estonian Research Council and the U.S. National Science Foundation.
This article was first published by the Penn State Eberly College of Science. Read the original.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Avoiding common figure errors in manuscript submissions
The three figure issues most often flagged during JBC’s data integrity review are background signal errors, image reuse and undeclared splicing errors. Learn how to avoid these and prevent mistakes that could impede publication.

Ragweed compound thwarts aggressive bladder and breast cancers
Scientists from the University of Michigan reveal the mechanism of action of ambrosin, a compound from ragweed, selectively attacks advanced bladder and breast cancer cells in cell-based models, highlighting its potential to treat advanced tumors.

Lipid-lowering therapies could help treat IBD
Genetic evidence shows that drugs that reduce cholesterol or triglyceride levels can either raise or lower inflammatory bowel disease risk by altering gut microbes and immune signaling.

Key regulator of cholesterol protects against Alzheimer’s disease
A new study identifies oxysterol-binding protein-related protein 6 as a central controller of brain cholesterol balance, with protective effects against Alzheimer’s-related neurodegeneration.

From humble beginnings to unlocking lysosomal secrets
Monther Abu–Remaileh will receive the ASBMB’s 2026 Walter A. Shaw Young Investigator Award in Lipid Research at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

Chemistry meets biology to thwart parasites
Margaret Phillips will receive the Alice and C. C. Wang Award in Molecular Parasitology at the ASBMB Annual Meeting, March 7-10 in Washington, D.C.

