Anne-Claude Gingras:
probing phosphatase complexes
There are two sides to every story, two halves that make a whole. In signal transduction, the cascade of protein activity that converts extracellular signals into an intracellular response, the complementary parts of the tale are the protein kinases and phosphatases – the complementary enzymes that drive signal transduction by adding and removing phosphate groups on target proteins.
 Rising science star Anne-Claude Gingras uses proteomics and other approaches to understand phosphatase biology and cell signaling networks.
Rising science star Anne-Claude Gingras uses proteomics and other approaches to understand phosphatase biology and cell signaling networks.
As a graduate student in the mid 1990s, Anne-Claude Gingras was struck by the contrast between knowledge of these two enzymes. "We knew so much more about the kinase side of the story than about the phosphatases," she says. "That didn't seem right, so I wanted to investigate phosphatases further."
Now a senior investigator at the Samuel Lunenfeld Research Institute at Mount Sinai Hospital in Toronto as well as an associate professor in the department of molecular genetics at the University of Toronto, Gingras has done just that – and more.
Not yet 40 years old, Gingras rapidly is establishing herself as an international leader in understanding how phosphatases are regulated and how they recognize their substrates. She's also become an innovator in the proteomics field, having developed experimental and computational tools that can better characterize dynamic protein-protein interactions.
It's quite a trajectory for someone who just 17 years ago was a bright but self-professed naïve graduate student who could barely speak or write English. But it's a path that anyone who has worked with Gingras will say is well deserved.
Cookbooks and circuitry
The first monumental step in this research journey occurred in 1994, when Gingras set foot at McGill University to begin graduate school. Though only a little more than 150 miles from her hometown on the outskirts of Québec city, the vibrant multicultural metropolis of Montreal seemed like another country to the young Québécois.
"I had never really left home before," she says, "but I knew I had to broaden my horizons, and learn English, if I could fully realize my potential. I thought that going to school in Montreal would be a great compromise. I could do research in English at McGill yet order food in French and therefore not die of starvation in my first year."
And while the first few months were challenging, her talents shone through fairly quickly. For while Gingras may not have been worldly, she did possess a great deal of scientific know-how.
For starters, her parents, both nurses, had fostered Gingras' creative and intellectual spirit since early childhood. "My father spent his spare time working on electronics, and I used to love helping him install transistors on circuit boards," she recalls. "He also had a fascination with chemistry and aerospace – my grandmother told me some pretty entertaining stories relating to his improper storage of rocket fuel – and hoped that one of the kids would become an astronaut or a chemist. I ended up the closest."
Gingras' mother, meanwhile, helped her develop her creative side and her lab hands by teaching her crafts and cooking. "She also bought me this super cool book of experiments called 'Le Petit Débrouillard' ('The Small Resourceful One'). I did every experiment in the book, some more than once."
After high school, Gingras stayed close to home and attended Laval University, where she studied biochemistry. The program at Laval focused heavily on laboratory and experimental work, and, by the time she graduated, Gingras had already developed significant hands-on experience.
"However, what really convinced me to go to graduate school was my summer project working for a just-starting young professor, André Darveau. As a new appointee, Darveau had a small team and taught me several important techniques himself while giving me a lot of independence. I really caught the research bug!"
Darveau also helped Gingras select some potential graduate advisors to meet with at McGill, which eventually led to her accepting a position with Nahum Sonenberg, whose group studied the mechanisms that control protein translation.
A productive start
Right as Gingras joined the lab, another graduate student, Arnim Pause, had just cloned two insulin-regulated small proteins that bind to eukaryotic translation initiation factor 4E, the protein that binds the cap on the end of messenger RNA and brings the RNA to ribosomes. These two peptides, 4E-BP1 and 4E-BP2, were found to inhibit cap-dependent translation when they bound.
"The topic was fresh and exciting, and I was able to work on a project that became a nexus for all of the lab's interests," she says. "I could contribute to projects ranging from structural biology to virology to mouse development."
The work cemented an interest in how protein phosphorylation regulates protein activity and also set off a graduate training career of which even the most demanding taskmaster would be proud. The various projects she collaborated on at McGill resulted in nearly 50 scientific publications, a number that continues to grow to this day.
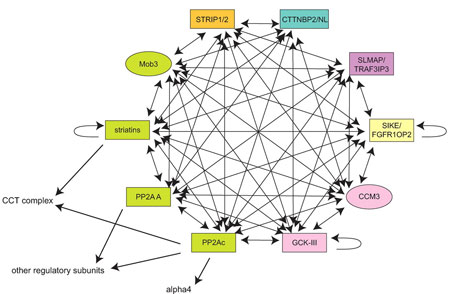 The Highly Interactive STRIPAK Complex. Model of the numerous protein-protein interactions (including self-interactions) within the STRIPAK complex, as unraveled by Gingras and her colleagues. Green nodes represent the previously characterized association between PP2Ac, PP2A A, striatins, and Mob3; pink nodes indicate previously characterized interactions between CCM3 and the CGK-III subgroup. From Mol. Cell. Proteomics8, 157-171.
The Highly Interactive STRIPAK Complex. Model of the numerous protein-protein interactions (including self-interactions) within the STRIPAK complex, as unraveled by Gingras and her colleagues. Green nodes represent the previously characterized association between PP2Ac, PP2A A, striatins, and Mob3; pink nodes indicate previously characterized interactions between CCM3 and the CGK-III subgroup. From Mol. Cell. Proteomics8, 157-171.
Gingras is quick to point out she owes a good deal of that remarkable number to arriving at Sonenberg's lab at an extremely opportune time and that most of the work was collaborative.
In addition, she spent a little longer than average in her graduate lab as she waited for her partner, Brian Raught (now a researcher at the Ontario Cancer Institute), so they could move on together. "That worked out really well," she notes. "I had enough data to graduate, but I knew once I started my postdoc the pressure to get positive findings would start again. But in this intermediate period, I had this window where I could try these fun, fishing-type projects, and I took full advantage of that opportunity."
One of Gingras' favorite studies during her McGill time was a project that combined old-school gel electrophoresis with mass spectrometry to identify 4E-BP1 phosphorylation sites regulated by the mTOR signaling pathway, which then led to identifying the in vivo hierarchy of 4E-BP1 phosphorylation. That publication played a big part in stimulating a desire to do postdoctoral training in proteomics.
Fortuitously, one of the best proteome researchers, Ruedi Aebersold at the Institute for Systems Biology in Seattle, was a long-time collaborator of Sonenberg.
"Ruedi had just published his first ICAT paper, which documented the use of stable isotopes to quantify proteins," she says. "I had already collaborated extensively with Ruedi's group, particularly Steve Gygi, who is now at Harvard, to identify phosphorylation sites on various translation factors, and I thought that was the place to go to learn how to perform quantitative proteomics and phosphoproteomics."
La grande débrouillarde
In 2005, after her three-year stint in Aebersold's lab, Gingras found herself a wanted woman. Research institutes from across the globe were offering jobs to the rising star. In the end, she decided to come back close to home and joined the Samuel Lunenfeld Research Institute in Toronto.
"Having spent a little time in Canada," she says jokingly, "I knew that Toronto was a superb environment for signaling biology. But during the interview process, what also became apparent was the collaborative spirit that ran through the Toronto community."
"Researchers here work together whenever it makes sense, which provides an extremely stimulating research environment both for faculty and trainees. And given the type and scope of research that I wanted to perform, robust collaborations were essential."
Gingras believes this human element cannot be understated. Even more than her many published papers, Gingras values the many great scientists she has had a chance to work with over the years and the opportunities those interactions opened up.
"The research world is smaller than you think," she says, "and the people you meet along the way will probably come back into your life at some point. So, for young scientists out there, when someone requests some cells or protein from you, be as nice as possible, because you never know what may happen in the future."
As for the present, Gingras' group combines interaction proteomics, phosphoproteomics, functional screens and other approaches to identify and characterize protein phosphatase complexes, several of which she initially identified through discovery proteomics. One such phosphatase complex of interest, which Gingras first discovered while working under Aebersold, is PP4cs, which is linked to DNA damage repair and also is associated with resistance to the potent cancer drug cisplatin.
Another current project, and the subject of two recent Journal of Biological Chemistry articles, is a large protein complex termed striatin interacting phosphatase and kinase, or STRIPAK. This complex contains both a phosphatase and a kinase bridged by a protein called CCM3 (so named because when mutated, it causes blood vessel defects in the brain called cerebral cavernous malformations). Her group has found that STRIPAK is involved in polarizing Golgi in cells, and they are now further characterizing structure and function.
In the near future, however, Gingras hopes to take a broader approach and try to model signaling networks quantitatively, and she has begun doing some studies in yeast. "On the phosphatase front, we still need to carry out detailed biochemical analyses to understand how they are regulated and how they recognize their targets," she notes. "But at the same time, we need to look at the network level if we want to figure out all the implications of crosstalk and feedback loops."
What's the score?
"Proteomics has tremendously advanced, even in the few years since I arrived at Lunenfeld," Gingras says, "and it has been very exciting to watch this progress. But new possibilities also bring up new challenges, things like ensuring that data quality is kept high and that the data can be shared in an appropriate format with the whole scientific community."
Gingras believes specialized journals like Molecular and Cellular Proteomics, of which she is an editorial board member, have been pioneers in this respect, but more general journals still need to improve their mechanisms to ensure data quality and data sharing. "Funding agencies should also step up and help facilitate funding of projects that address these issues, such as maintaining data repositories."
Of the many challenges that fall under the umbrella of figuring out what to do with all the data, Gingras has most keenly been intrigued with data scoring.
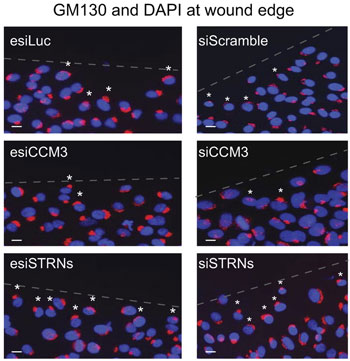 STRIPAK and the Golgi. Another of Gingras' recent discoveries with STRIPAK is its role in Golgi orientataion. Here, cells depleted of STRIPAK component CCM3 by esiRNA (left) or chemical siRNA (right) show reduced numbers of Golgi properly positioned (white asterisks) in response to a wound (shown by dashed line). esiLuc and siScramble are negative controls. From J. Biol. Chem.286, 25065-25075.
STRIPAK and the Golgi. Another of Gingras' recent discoveries with STRIPAK is its role in Golgi orientataion. Here, cells depleted of STRIPAK component CCM3 by esiRNA (left) or chemical siRNA (right) show reduced numbers of Golgi properly positioned (white asterisks) in response to a wound (shown by dashed line). esiLuc and siScramble are negative controls. From J. Biol. Chem.286, 25065-25075.
"It is a major issue in the field," she says. "Proteomics can identify all sorts of protein-protein interactions, but how can we be sure they are genuine? I hate to imagine a proteomics study producing a novel interaction, but later on a poor graduate loses weeks or months of work unsuccessfully doing biochemical follow-ups because the interaction was spurious."
To rescue frustrated students across the world, Gingras teamed up with colleague Alexey Nesvizhskii at the University of Michigan to develop an innovative computer program called SAINT (for Significance Analysis of Interactome); this groundbreaking probability-based program analyzes several metrics to quantify protein interactions in a data set as true or false.
The seeds for SAINT go back more than eight years, starting with some informal discussions and idea exchanges between Gingras and Nesvizhskii when they were both postdocs with Aebersold. "Back then, though, we didn't have the technology to realize our ideas, but a couple of years ago we reconnected and thought, you know, this could work now."
Gingras believes tools like SAINT will have tremendous impact on how researchers analyze, score and report their protein-protein interactions. With Mike Tyers at the University of Edinburgh in Scotland, Gingras has been working on a complementary program called ProHits that lets scientists store, search and analyze mass-spectrometry data. She now is working on interfacing ProHits and SAINTS to create a thorough software tool.
Since that first day in Sonenberg's lab, Gingras' life certainly has been a whirlwind of activity, but 17 years, more than 80 papers, and several novel discoveries later, she still is ready for more.
"The power of modern mass spectrometers for quantification is only beginning to be tapped into by systems biologists," she says. "In interaction proteomics, for example, most large-scale studies involve static interaction maps, which reveal nothing about protein complex assembly, the stoichiometry of the subunits or the dynamics of the interactions. Over the next few years, we will see all these quantitative measures implemented into large projects both in systems biology and clinical proteomics. It's going to be fun."
"Soon, I will no longer be considered a young researcher," she adds, "but in this field I can stay young at heart for a very long time."
References
Ceccarelli, D. F., Laister, R. C., Mulligan, V. K., Kean, M. J., Goudreault, M., Scott, I., Derry, W. B., Chakrabartty, A., Gingras, A. C., and Sicheri, F. (2011) CCM3/PDCD10 heterodimerizes with GCKIII kinases using a mechanism analogous to the CCM3 homodimerization mechanism.J. Biol. Chem. May 11. [Epub ahead of print]
Kean, M. J., Ceccarelli, D., Goudreault, M., Sanches, M., Tate, S., Larsen, B., Gibson, L. C., Derry, W. B., Scott, I. C., Pelletier, L., Baillie, G. S., Sicheri, F., and Gingras, A. C. (2011) Structure-function analysis of core STRIPAK: a signalling complex implicated in golgi polarization. J. Biol. Chem. May 11. [Epub ahead of print]
Choi, H., Larsen, B., Lin, Z. Y., Breitkreutz, A., Mellacheruvu, D., Fermin, D., Qin, Z. S., Tyers, M., Gingras, A. C., and Nesvizhskii, A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods8, 70 – 73.
Liu, G., Zhang, J., Larsen, B., Stark, C., Breitkreutz, A., Lin, Z. Y., Breitkreutz, B. J., Ding, Y., Colwill, K., Pasculescu, A., Pawson, T., Wrana, J. L., Nesvizhskii, A. I., Raught, B., Tyers, M., and Gingras, A. C. (2010) ProHits: integrated software for mass spectrometry-based interaction proteomics.Nat. Biotechnol.28, 1015 – 1017.
Gingras, A. C., Caballero, M., Zarske, M., Sanchez, A., Hazbun, T. R., Fields, S., Sonenberg, N., Hafen, E., Raught, B., and Aebersold, R. (2005) A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol. Cell. Proteomics4, 1725 – 1740.
Gingras, A. C., Raught, B., Gygi, S. P., Niedzwiecka, A., Miron, M., Burley, S. K., Polakiewicz, R. D., Wyslouch-Cieszynska, A., Aebersold, R., and Sonenberg, N. (2001) Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev.15, 2852 – 2864.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in People
People highlights or most popular articles

Decoding how bacteria flip host’s molecular switches
Kim Orth will receive the Earl and Thressa Stadtman Distinguished Scientists Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Thiam elected to EMBO
He was recognized during the EMBO Members’ Meeting in Heidelberg, Germany, in October.

The timekeepers of proteostasis
Learn about the cover of the winter 2026 ASBMB Today issue, illustrated by ASBMB member Megan Mitchem.

Defining JNKs: Targets for drug discovery
Roger Davis will receive the Bert and Natalie Vallee Award in Biomedical Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Building better tools to decipher the lipidome
Chemical engineer–turned–biophysicist Matthew Mitsche uses curiosity, coding and creativity to tackle lipid biology, uncovering PNPLA3’s role in fatty liver disease and advancing mass spectrometry tools for studying complex lipid systems.

Summer research spotlight
The 2025 Undergraduate Research Award recipients share results and insights from their lab experiences.

