‘How life began merits a preceding discussion of what life actually is’
The origin of life has perplexed and fascinated scientists for the better part of a century. Nir Ben–Tal and Amit Kessel’s new book, “From Molecules to Cells: The Origin of Life on Earth,” uses biochemistry and evolution to describe hypotheses of life’s origins and the last universal common ancestor, an anaerobic prokaryote.
According to Ben–Tal and Kessel, life likely began on Earth around 3.8 to 3.5 billion years ago with a combination of RNA–peptide–lipid–metal molecules. They said the two key operational capabilities to life are informational (encoding and transmitting genetic information) and metabolic (accelerating chemical processes and utilizing nutrients).
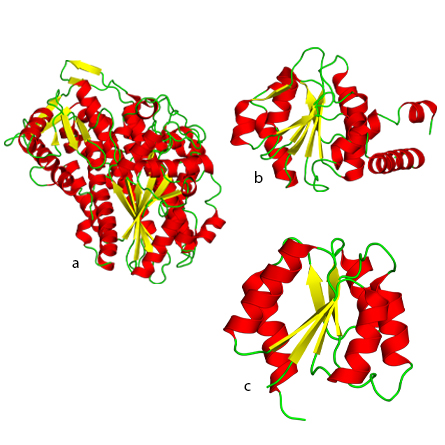
a. The P-loop fold of nucleoside triphosphate hydrolases.
b. NAD(P)-binding Rossmann fold.
c. Flavodoxinlike fold.
“From Molecules to Cells” focuses specifically on the biological macromolecules that are critical to life as we understand it, such as nucleic acids, proteins and lipids. The chapter on protein evolution describes the nine most ancient folds of protein domains, including the idea that most gene families formed from 3.3 to 2.85 billion years ago and new protein families formed mainly from existing genes after that time.
The book describes hypotheses and experiments that show how amino acids and simple molecules related to the cell can arise spontaneously, given the right conditions. Amino acids and similar molecules are found on meteorites, suggesting this chemistry also occurs on other planets.
This interview has been edited.
What motivated you to write this book?
Kessel: As biologists, we were mainly motivated by the need to bring more biology into the story. Most origin-of-life textbooks focus on the chemical evolution step of life’s emergence — the organic chemistry that spontaneously turned simple compounds into biomoleculelike proteins, RNA and peptides on primordial Earth. The nature of these biomolecules and how they evolved into functionally efficient forms capable of forming biochemical networks is usually ignored.
Today, it is possible to track some of these processes thanks to the ample biological data obtained in recent years on proteins and on the genes coding for them. This has allowed scientists to use a top-down approach to analyze the sequences and structures of contemporary proteins in order to trace their evolution back to early peptides and peptide–RNA complexes, which are similar to those created by the chemical evolution step.
Ben–Tal: Several years ago, I gave a talk about our discovery of protein segments that are extensively reused in the protein world, suggesting that they might be evolutionary descendants of ancient functional peptides.
The talk was meant mostly for undergraduate bioinformatics students, and I felt that I should briefly introduce them to the origin of life in order to give a broader context of our own research. The very enthusiastic response to this talk is what made us think about writing a book that integrates protein evolution with the preceding step of chemical evolution. Obviously, the book is based for the most part on studies by many other scientists, including many that are no longer with us, and our own research adds just a tiny bit.
We also felt that the discussion of how life began merits a preceding discussion of what life actually is. To this end, we included a chapter that describes the basic features that are common to all living organisms on Earth. We then used these features to compare the general definitions of life used widely today, such as that used by NASA, with a narrower yet more accurate definition that refers to life as we know it on our planet.
Did “From Molecules to Cells” stem from your interest in protein evolution?
Ben–Tal: In our lab, we exploit evolutionary data, on the one hand, to highlight key aspects of a protein of interest, as in ConSurf, and, on the other hand, to infer from detailed mechanistic studies about one protein into homologues. In general, the ample sequence and structure data enables investigation of proteins as families.

In recent years, we discovered numerous protein fragments, which we call “themes,” that are extensively reused by proteins. This discovery, followed by the finding that some of these themes are involved in ancient protein functions like the binding of nucleotide cofactors, suggested that they may be descendants of peptides that preceded proteins.
The book is our effort to bridge the gap between what is known from the bottom-up chemical evolution approach and what we are beginning to reveal from peptide/protein evolution.
Elaborate a bit on what you call the world hypotheses, including the RNA world, the peptide/amyloid world, the lipid world and the virus world.
Kessel: The world theories came up when scientists looked for a molecule that could serve as a progenitor of life. Such a molecule would have to be simple enough to form spontaneously under primordial Earth conditions yet sufficiently sophisticated to act in both informational and operational capacities. That is, to be able to hold genetic information, pass it on to the next generations, promote biochemical processes and evolve.
In all the living organisms we know, the informational role is carried out by DNA and the operational role is carried out by proteins and RNA, and they are all coupled via the gene expression system. This could not be the case in the first protocells, because such complex systems did not exist. So the same molecule needed to function in both informational and operational roles.
DNA and proteins were excluded because DNA cannot catalyze processes and proteins are too complex to form spontaneously. Walter Gilbert proposed his RNA world theory when he and others realized that RNA molecules, already known to store genetic information, catalyze chemical reactions in contemporary organisms.
Other researchers later proposed similar world theories for peptides (isolated or in the form of stable amyloid structures) and for lipids, which were also found to be catalytically active and form spontaneously. These theories were spiced with proposed hybrid molecules such as ‘peptide nucleic acid,’ which has stable peptide backbone and nucleotide side chains that can form base pairs.
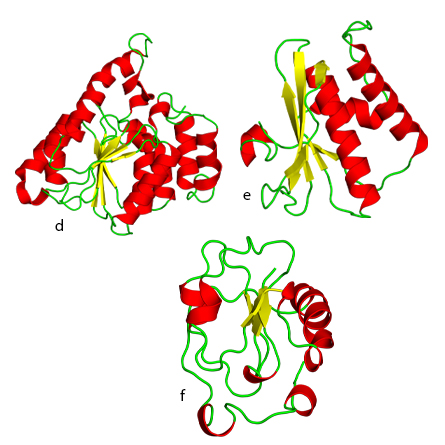
e. Ribonuclease H-like fold.
f. Ferredoxinlike fold.
Ben–Tel: Although the RNA world seems to be the most plausible theory, it is burdened by certain aspects of RNA molecules, such as their unstable nature and very limited catalytic activity. Moreover, to evolve, RNA molecules would have to self-replicate by template-based synthesis. Such a process has been demonstrated only for short or otherwise simple RNA molecules, which cannot act as catalytic entities.
While these problems remain unresolved, some of them could be mitigated if the RNA molecules formed molecular complexes with peptides, metals and lipid molecules (isolated or as assemblies). Binding any of these entities would stabilize the RNA molecule by suppressing internal dynamics and masking charges.
And each of these entities could contribute its unique advantages: Peptides and metals could mediate substrate binding and increase the catalytic efficiency of the RNA, whereas peptides and lipids could increase the catalytic scope of the RNA. In our view, such multichemical entities’ synergy was crucial for completing the emergence-of-life process.
What do you envision when you think of Earth at the time of the last universal common ancestor?
Kessel: Life is believed to have appeared during the early Archaean eon. At this time, Earth has already cooled down, the molten rock on its surface solidified and water vapor condensed into oceans. It’s difficult to know exactly how the planet looked back then, because very little geological evidence is left.
We envision this Earth as covered largely by water, with considerable volcanic and hydrothermal activity. This is in line with the widely accepted view of life’s emergence on the planet; the process is thought to have occurred in hydrothermal microenvironments, which could provide all that was required for the chemical evolution stage — starting materials, an energy source (heat) and even mineral catalysts. Most of these microenvironments were probably hydrothermal vents on the oceans’ floors, whereas others were on land, such as hot springs and geysers. The latter two were probably more important for the chemical evolution stage, since the air–water–land interfaces they contained promoted polymerization reactions. The process was affected by the atmosphere, which contained mainly nitrogen and carbon dioxide but also several greenhouse gasses. The latter kept the planet relatively warm despite the faint sun.
Building off this idea, briefly discuss how life arose on this planet.
Ben–Tal: We assume the process included the gradual formation of increasingly complex organic molecules from simple compounds like methane, ammonia, hydrogen cyanide and hydrogen sulfide via spontaneous chemical reactions. This ultimately led to the formation of biomolecular building blocks that polymerized to form functional peptides and RNA molecules.
The first clue that such a process was possible came from the Miller–Urey experiment in 1953, which demonstrated the spontaneous formation of amino acids in a closed system containing only methane, ammonia, hydrogen and water vapor subjected to electric sparks. Indeed, amino acids are extremely easy to form and can be found also inside meteorites.
Kessel: Some of these molecules are thought to have been sufficiently complex to carry out simple life processes, and when they became trapped inside lipid vesicles along with their substrates, they formed the first protocells. Continued evolution of the latter into real cells was accompanied by morphological and functional changes as well as the development of a DNA genome, a protein translation system and a protein-catalyzed metabolism.
What about life on other planets?
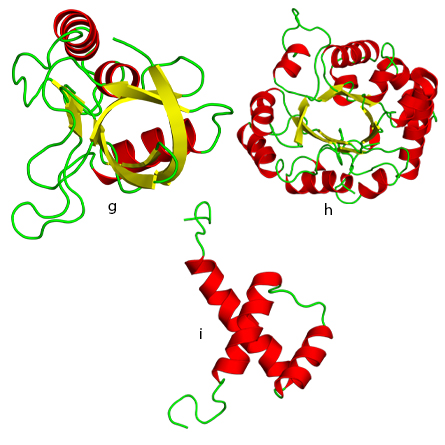
h. TIM beta/alpha barrel fold.
i. DNA/RNA-binding 3-helical bundle fold.
Kessel: Could such a process happen on other planets as well? I don’t see why not. There are a lot of planets out there and, given a set of starting materials, an energy source and liquid water, we have no reason to believe that complex molecules could not form as they did on Earth and form simple living organisms.
In our solar system, a few candidates are proposed. For example, Mars is relatively close to the sun for biological organisms that may live there to harness solar energy, and it also contains subsurface liquid water. Still, these organisms would have to cope with the strong radiation on the planet’s surface.
Europa and Enceladus, moons of Jupiter and Saturn, respectively, are very far from the sun, and their surfaces are completely icy. However, below their surfaces, there are high-temperature activities that might promote life the same way hydrothermal vents are thought to have done on primordial Earth.
How do you think proteins evolved into the complex molecular machines we see in extant organisms?
Kessel: This is one of the toughest questions to answer; while there is ample evidence about the major mechanisms of protein evolution — which involve replication, fission, divergence and recombination of domains and smaller parts — it is very difficult to know exactly how a certain protein or molecular machine evolved. This is because we rarely have enough representatives of the intermediate states of the process, especially when it is very old.
Ben–Tal: The ribosomal complex is one case where information obtained from numerous phylogenetic studies was used to suggest an almost complete evolutionary process, although these studies focused on the RNA components of the complex. The process started from catalytic RNAs capable of randomly attaching amino acids, continued in a separate development of a decoding center that bound amino acid–carrying tRNA, and culminated in the assembly of the two parts into a genetically guided protein-synthesizing machine.
Some phylogenetic studies try to date the evolutionary processes of proteins by assigning the appearance of domains and folds to known evolutionary and geological events, such as the emergence of eukaryotes and multicellular organisms and the oxidation of the atmosphere. This yielded the proposed time frame for the appearance of the first protein gene families. Such time calibrations of processes are generally inaccurate and should be taken with caution.
Kessel: Still, even a qualitative investigation of protein evolutionary processes yields very interesting findings. For example, the stepwise evolution of different protein functions, starting with redox catalysis and nucleotide/iron–sulfur binding, followed by respiration, photosynthesis and signaling. It is also interesting to see how the first protein folds that appeared match these ancient functions, such as nucleotide binding by the Rossmann, P-loop and flavodoxin folds.
About the authors

Nir Ben–Tal is a professor of biochemistry and molecular biology and heads the Ben–Tal Lab of Computational Structural Biology at Tel Aviv University. He received his Ph.D. from the Technion–Israel Institute of Technology and conducted postdoctoral research at Columbia University. He is an author on more than 150 publications and has trained more than 50 graduate students and postdoctoral fellows. His lab created the ConSurf server; the 2010 and 2016 papers describing this resource have been cited over 3,000 times. He is senior editor of the journal Protein Science and a member of the editorial boards of the scientific journals PLoS Computational Biology, eLIFE and the Journal of Biological Chemistry.

Amit Kessel is a research associate at Tel Aviv University, where he works as a computational biologist and a lecturer. He received a Ph.D. from Tel Aviv University and conducted postdoctoral research at Columbia University. His website provides a summary of the new book, describes his other work, and provides general resources for scientists, students and interested members of the general public. Follow him on Twitter: @KesselAmit.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Opinions
Opinions highlights or most popular articles

Women’s health cannot leave rare diseases behind
A physician living with lymphangioleiomyomatosis and a basic scientist explain why patient-driven, trial-ready research is essential to turning momentum into meaningful progress.

Making my spicy brain work for me
Researcher Reid Blanchett reflects on her journey navigating mental health struggles through graduate school. She found a new path in bioinformatics, proving that science can be flexible, forgiving and full of second chances.

The tortoise wins: How slowing down saved my Ph.D.
Graduate student Amy Bounds reflects on how slowing down in the lab not only improved her relationship with work but also made her a more productive scientist.

How pediatric cataracts shaped my scientific journey
Undergraduate student Grace Jones shares how she transformed her childhood cataract diagnosis into a scientific purpose. She explores how biochemistry can bring a clearer vision to others, and how personal history can shape discovery.

Debugging my code and teaching with ChatGPT
AI tools like ChatGPT have changed the way an assistant professor teaches and does research. But, he asserts that real growth still comes from struggle, and educators must help students use AI wisely — as scaffolds, not shortcuts.

AI in the lab: The power of smarter questions
An assistant professor discusses AI's evolution from a buzzword to a trusted research partner. It helps streamline reviews, troubleshoot code, save time and spark ideas, but its success relies on combining AI with expertise and critical thinking.

