From the journals: MCP
Muscling in on a signaling pathway. Probing weaknesses in the T cell surface. Improving single-cell proteomics two ways. Read about papers on these topics recently published in Molecular & Cellular Proteomics.
Muscling in on a signaling pathway
Muscle-specific receptor tyrosine kinase, or MuSK, plays an essential role in forming neuromuscular junctions. It is activated by a nerve-derived proteoglycan called agrin through low-density lipoprotein receptor–related protein 4, or LRP4. Several neuromuscular diseases are associated with the MuSK signaling pathway; in one of these, myasthenia gravis, autoantibodies target various components of that pathway. Similarly, mutations in genes that function within the pathway characterize congenital myasthenic syndromes.
Researchers use MuSK-activating antibody, an agonist antibody, or Ab, to explore the benefits of MuSK activation at the neuromuscular junction. In two recent studies, this Ab acted in an LRP4-independent manner in mouse models of amyotrophic lateral sclerosis. Both studies reported delayed denervation upon treatment with the MuSK agonist Ab, with notable differences in downstream functional outcomes. Given these observations, Hanna Budayeva and a team of researchers at Genentech used Ab to compare direct MuSK activation to coreceptor sequestration and assess how much Ab-mediated MuSK activation recapitulates the physiological agrin-mediated activation.
The team performed dose–response and time-course experiments on C2C12 myotubes and systematically compared site-specific phosphorylation downstream of MuSK activation by agrin and MuSK-activating Ab. Using mass spectrometry–based proteomics, they tested two MuSK agonists, and observed novel but similar intracellular responses at known and newly identified MuSK signaling components. Among these responses was inducible tyrosine phosphorylation of multiple Rab GTPases that was blocked by MuSK inhibition. Moreover, mutation of this site in Rab10 disrupted association with its interacting proteins.
The researchers characterized MuSK signaling in depth and identified a role for Rab GTPases in the downstream signaling of this MuSK pathway. These findings were recently published in the journal Molecular & Cellular Proteomics.
Probing weaknesses in the T cell surface
T cells are white blood cells that play a central role in the body’s adaptive immune response. In particular, cytotoxic T cells, or CD8+ T cells, promote tumor-cell killing and are used in cancer therapy.
Researchers know the cell surface proteome, or surfaceome, mediates T cell interactions with the external environment but do not yet fully understand how external factors affect that surfaceome. Immunosuppressive factors in the tumor microenvironment, or TME, can impair T cell function and limit the antitumor immune response by changing cell surface receptors and surface proteins.
To study surface proteome remodeling in T cells in greater detail, James Byrnes and a research team at the University of California, San Francisco, performed quantitative cell surface proteomics on primary human CD8+ T cells to determine how immunosuppressive TME factors, Tregs (regulatory T cells a specialized subpopulation of T cells that act to suppress immune response), and hypoxia affected the activated CD8+ surfaceome. Consistent with suppressed cell activation, the Tregs partly reversed activation-induced changes in CD8+ T cells. Hypoxia triggered large-scale surfaceomic remodeling consistent with a general cellular response to the metabolic demands of a low-oxygen environment. In both conditions, the researchers saw reduced expression of proteins primarily involved in nutrient transport. These changes could limit T cell nutrient uptake during activation and, subsequently, T cell proliferation.
These findings, recently published in the journal Molecular & Cellular Proteomics, provide insight into the plasticity of the T cell surfaceome and lay the foundation for future efforts to characterize further and therapeutically engage T cell surface proteins for better immune response.
Improving single-cell proteomics two ways
Single-cell proteomics, or scMS, uses liquid chromatography–mass spectrometry based proteomics. As this technology evolves, researchers need to better understand the proteome, both the proteins quantified per cell and the quantitative performance thereof. However, multiplexing scMS is challenging; high ion injection times and high-resolution spectra are needed to quantify the single-cell signal.
A new real-time search, or RTS, on the Orbitrap Eclipse Tribrid mass spectrometer combined with synchronous precursor selection–triple-stage mass spectrometry, or SPS-MS3, acquisition could help researchers measure samples that are multiplexed using isobaric tags. To show this, Benjamin Furtwängler and a team of researchers from the University of Copenhagen, the Technical University of Denmark and other institutions compared classical MS2 acquisition to RTS-MS3 and a new RTS-MS2 acquisition method called RETICLE and found that both RTS methods outperformed classical MS2 acquisition.
The researchers recently published their findings in the journal Molecular & Cellular Proteomics. They found that scMS data acquisition using the tribrid design in combination with RTS can result in better scMS data sets. RTS-MS3 provided similar proteome coverage to MS2 at higher quantitative accuracy, whereas RETICLE resulted in higher proteome coverage. Thus, RETICLE could be especially useful for high-throughput applications, such as building single-cell atlases, whereas RTS-MS3 could provide the accuracy needed to model complex protein networks.
Enjoy reading ASBMB Today?
Become a member to receive the print edition monthly and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Genetics studies have a diversity problem that researchers struggle to fix
Researchers in South Carolina are trying to build a DNA database to better understand how genetics affects health risks. But they’re struggling to recruit enough Black participants.
Scientists identify new function of learning and memory gene common to all mammalian brain cells
Findings in mice may steer search for therapies to treat brain developmental disorders in children with SYNGAP1 gene mutations.
From the journals: JBC
Biased agonism of an immune receptor. A profile of missense mutations. Cartilage affects tissue aging. Read about these recent papers.
Cows offer clues to treat human infertility
Decoding the bovine reproductive cycle may help increase the success of human IVF treatments.
Immune cells can adapt to invading pathogens
A team of bioengineers studies how T cells decide whether to fight now or prepare for the next battle.
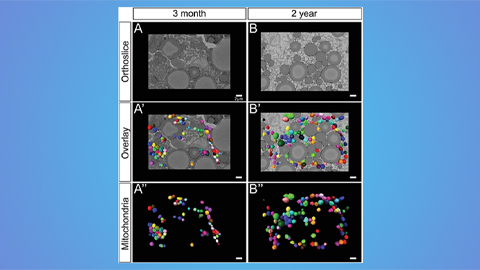
Hinton lab maps structure of mitochondria at different life stages
An international team determines the differences in the 3D morphology of mitochondria and cristae, their inner membrane folds, in brown adipose tissue.

