From the journals: MCP
A triphosphorylated species may be linked to DNA repair. A site-specific proteomics approach holds promise for Alzheimer’s diagnosis. A new platform improves quantification of histone modifications. Read about papers on these topics recently published in Molecular & Cellular Proteomics.
A lipid in the nucleus
Phosphatidylinositol is a complex molecule. The lipid’s head group, a sugar, has three potential phosphorylation sites, generating seven potential phosphorylated species called polyphosphoinositides, each of which is thought to bind a different set of effector proteins and exert diverse effects.
Researchers long have known that cleavage of the head group from its hydrophobic fatty acid tails forms a cytoplasmic second messenger. More recent research has identified polyphosphoinositides, complete with fatty acids, in nonmembrane environments. Researchers have mapped the interactions of a few phosphoinositide species with cytoplasmic and nuclear proteins, but they do not understand yet the role of the triphosphorylated species PIP3.
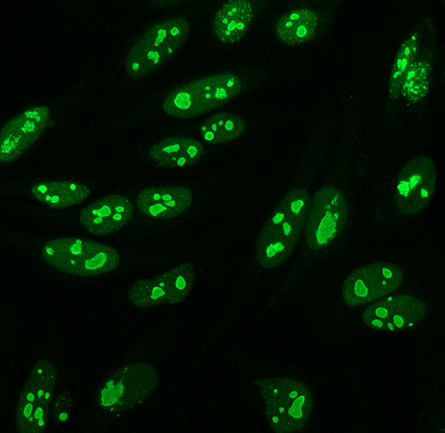
In a recent article in the journal Molecular & Cellular Proteomics, postdoctoral researcher Fatemeh Mazloumi Gavgani and colleagues at the University of Bergen in Norway describe how they used immunofluorescence to determine that nuclear PIP3 mostly is located in the nucleolus, where ribosomes are assembled. Then they used mass spectrometry to identify PIP3-interacting proteins from the nuclear fraction of HeLa cells; most of the proteins were nucleolus residents with known roles in RNA processing, splicing or catabolism, or translation.
One binding partner was poly(ADP-ribose) polymerase 1, or PARP1, an enzyme that adds a chain of ADP-ribosyl subunits to protein substrates after DNA damage, mobilizing proteins involved in damage repair. Through truncation analysis and directed mutations, the authors found that three polybasic regions in PARP1 rich in arginine and lysine residues mediate its binding to PIP3. The authors argue that the interaction suggests a link between PIP3 and DNA repair that merits further study.
Given that the bulk of each PIP3 molecule is hydrophobic, the researchers still aren’t sure how it might be transported to or synthesized in the nucleolus and remain in solution. They speculate that the lipid could form micelles or be shielded by binding to carrier proteins.
New approach holds promise for Alzheimer’s diagnosis
The cerebrospinal fluid, or CSF, that surrounds and supports the central nervous system, or CNS, plays an important role in brain development and neuronal functioning. It is the only body fluid that directly interchanges with the extracellular fluid of the CNS and reflects ongoing pathological changes in the CNS. Thus, proteomic analysis of CSF has great potential for CNS-related disease diagnostics.
Glycosylation, an important post-translational modification, regulates many cellular processes. Defects in glycosylation have been linked to human diseases, making it a valuable target to study. While researchers have done extensive proteome profiling for CSF, few studies aim at unraveling a site-specific CSF N-glycoproteome.
In their recent paper in Molecular & Cellular Proteomics, Zhengwei Chen, Qinying Yu and a team at the University of Wisconsin–Madison describe their large-scale site-specific approach for in-depth CSF N-glycoproteome analysis, which allows for thorough coverage of CSF N-glycopeptides, including glycopeptide sequences, glycosylation site and glycan composition. In addition to generating the largest reported N-glycoproteome data set for CSF to date, the researchers applied this strategy to compare the N-glycoproteome coverage between people with and without Alzheimer’s disease. They uncovered diverse and distinct glycosylation patterns, paving the way for promising glycosylation-based biomarker candidates for Alzheimer’s. This work lays a foundation for more in-depth investigation of the functional roles of these glycosylated proteins in the progression of Alzheimer’s and other neurodegenerative diseases.
New platform to quantify histone methylation
Histone modifications are post-translational modifications to histone proteins that affect gene expression by altering chromatin structure and are responsible for packaging long DNA molecules into more compact, denser structures. Methylation is a common histone modification mediated by the addition of one or more methyl groups on the lysine or arginine amino acid of the histone protein.
Methylation is important in gene regulation and has been linked to progression of many cancers; however, researchers have difficulty quantifying this histone modification because of the spatial complexities that result when more than one methyl group is added on the same amino acid.
Francesca Zappacosta and a team of researchers at GlaxoSmithKline recently developed a simplified mass spectrometry–based platform for histone methylation analysis. The technique uses chemical derivatization to reduce the complexity of the protein to be analyzed, improving sensitivity for the detection of methylation. In one tested approach, the researchers quantified and distinguished between symmetrical and asymmetrical dimethylation of histone H4R3. They also were able to quantify modification levels as low as 0.02%. The team describes their mass spectrometric platform and findings in a recent article in Molecular & Cellular Proteomics.
This effective and robust MS-based approach holds promise for many studies that involve quantification of histone methylation profiles.
Enjoy reading ASBMB Today?
Become a member to receive the print edition monthly and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Genetics studies have a diversity problem that researchers struggle to fix
Researchers in South Carolina are trying to build a DNA database to better understand how genetics affects health risks. But they’re struggling to recruit enough Black participants.
Scientists identify new function of learning and memory gene common to all mammalian brain cells
Findings in mice may steer search for therapies to treat brain developmental disorders in children with SYNGAP1 gene mutations.
From the journals: JBC
Biased agonism of an immune receptor. A profile of missense mutations. Cartilage affects tissue aging. Read about these recent papers.
Cows offer clues to treat human infertility
Decoding the bovine reproductive cycle may help increase the success of human IVF treatments.
Immune cells can adapt to invading pathogens
A team of bioengineers studies how T cells decide whether to fight now or prepare for the next battle.
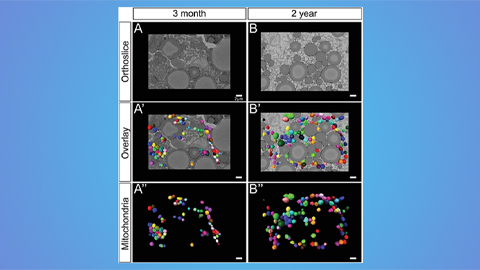
Hinton lab maps structure of mitochondria at different life stages
An international team determines the differences in the 3D morphology of mitochondria and cristae, their inner membrane folds, in brown adipose tissue.


