From the journals: MCP
Three ways of looking at mass spectrometry: analyzing secreted proteins, imaging mass spectrometry for clinical use and spectral libraries for MS data analysis. Read about these papers recently published in the journal Molecular & Cellular Proteomics.
Analyzing secreted proteins
Cells communicate via signals in response to environmental changes. Proteins can act as signaling cues, and proteins released into the extracellular space are known as the secretome. Secreted proteins play an important role in many diseases, such as cancer and cardiovascular diseases, and are potential targets for drugs and diagnostics. Researchers study the secretome using liquid chromatography-mass spectrometry, or LC-MS, or secretomics. LC separates compounds within a sample, while MS measures the mass-to-charge ratio of molecules present in compounds, which helps quantify and identify proteins.
In a recent review paper in Molecular & Cellular Proteomics, Sascha Knecht and a team from GlaxoSmithKline and the Technical University of Munich explored the challenges, approaches and applications of LC-MS–based secretomics. They noted that obstacles to achieving accurate results include cell death, serum proteins in cell culture studies, posttranslational modifications, unconventional secretion events and a high background signal due to intracellular proteins.
The three approaches to LC-MS-based secretomics are serum-free secretomics, metabolic labeling and proximity labeling. In serum-free secretomics, proteins are directly quantified from serum-free cell culture supernatants. In metabolic labeling, secreted proteins are labeled with affinity tags to selectively separate them from serum proteins. In proximity labeling, engineered enzymes are used to tag and enrich secreted proteins.
The authors explained that each approach has been applied in various biomedical research studies. For example, serum-free secretomics analysis has been used to study secreted proteins in macrophages, cellular communication in the immune system and cell death modes that contribute to inflammatory diseases. Metabolic labeling has been used to identify proinflammatory and immunomodulatory proteins as well as adipocytes. Proximity labeling has been used to track protein secretion in mouse blood plasma.
In summary, the authors stated that LC-MS–based secretomics gives an unbiased and comprehensive view of the secretome. With continued advancement, the authors noted that it may become a standard tool for studying secreted proteins in health and disease and ultimately in therapeutics.

Imaging mass spectrometry for clinical use
Imaging mass spectrometry, or IMS, is a technology that combines mass spectrometry with spatial analysis to provide an unbiased, highly specific 2D map of the molecules that make up tissues. In a recent paper in Molecular & Cellular Proteomics, Jessica L. Moore and a team of researchers at Vanderbilt University reviewed the potential of matrix-assisted laser desorption ionization IMS, a subtype of IMS, for clinical diagnostics and prognostics.
IMS allows researchers to probe tissue directly without the use of costly antibodies, and recent advances have significantly reduced the time needed for experiments. The authors explained that combining IMS with microscopy, which has high spatial resolution, enhances molecular visualization, increases mapping accuracy and gives IMS even greater potential as a standard pathology tool. However, clinical IMS assays often require bioinformatic analysis. Therefore, the authors noted that the technical knowledge needed for analysis as well as the government approval process for clinical use, which hinges on proof of valid and efficient assay performance, may delay clinical adoption.
Finally, the authors stated that IMS is a useful tool for studying the molecular makeup of diseases. With continued IMS advancements, this technology could become better suited for clinical applications such as diagnostics.
Spectral libraries for MS data analysis
As molecules pass through a mass spectrometer, energetically unstable ions are separated, or fragmented, and the resulting fragmentation patterns are reflected in the mass spectral data, which are useful for protein identification. Unlike conventional MS data analysis, which selectively fragments the most intense peptide ions, data-independent acquisition, or DIA, MS fragments all ions within a specific range of mass-to-charge ratios. Therefore, DIA MS is highly reproducible, reliable and accurate. However, complex processing of the DIA MS data may complicate analysis.
Two main approaches can address this challenge: library-based and library-free DIA MS. Library-based approaches analyze DIA MS data and assess proteins using a preconstructed spectral library that contains relative signal intensities of peptide fragment ions; this library either is built from prior conventionally analyzed data or predicted from peptide precursor sequences. Library-free approaches analyze data using a protein sequence database or predicted library. Different software tools cater to each type of analysis.
In a recent study in Molecular & Cellular Proteomics, Fangfei Zhang and a team at Westlake University and Westlake Omics in China evaluated the performance of five widely used types of DIA MS analysis software, using six data sets obtained from three different instruments. The authors used both library-based and library-free DIA MS to analyze each data set. They found that when the preconstructed spectral library was small and contained few proteins, library-based methods performed poorly and library-free methods provided more accurate and fruitful analyses. However, when the prebuilt spectral libraries were large and included a wider range of protein groups, library-based methods performed much better and were more accurate. This underscores the importance of constructing a comprehensive library for DIA MS analysis.
Enjoy reading ASBMB Today?
Become a member to receive the print edition monthly and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Genetics studies have a diversity problem that researchers struggle to fix
Researchers in South Carolina are trying to build a DNA database to better understand how genetics affects health risks. But they’re struggling to recruit enough Black participants.
Scientists identify new function of learning and memory gene common to all mammalian brain cells
Findings in mice may steer search for therapies to treat brain developmental disorders in children with SYNGAP1 gene mutations.
From the journals: JBC
Biased agonism of an immune receptor. A profile of missense mutations. Cartilage affects tissue aging. Read about these recent papers.
Cows offer clues to treat human infertility
Decoding the bovine reproductive cycle may help increase the success of human IVF treatments.
Immune cells can adapt to invading pathogens
A team of bioengineers studies how T cells decide whether to fight now or prepare for the next battle.
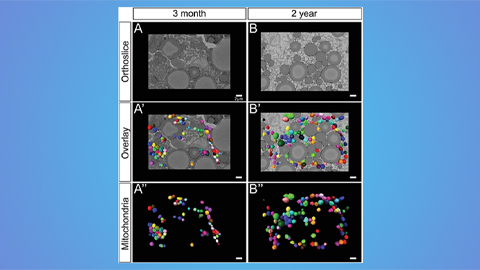
Hinton lab maps structure of mitochondria at different life stages
An international team determines the differences in the 3D morphology of mitochondria and cristae, their inner membrane folds, in brown adipose tissue.

