Scientists sweep cellular neighborhoods where Zika hides out
Most people infected with Zika never show symptoms. But the virus sometimes causes severe disability — from microcephaly in babies to weakness or partial paralysis in adults — and there is no treatment. In a paper in the journal Molecular & Cellular Proteomics, researchers report a comprehensive study of how the virus interacts with host cells. One of their findings gives insight into how Zika escapes immune signaling and proliferates inside the body.
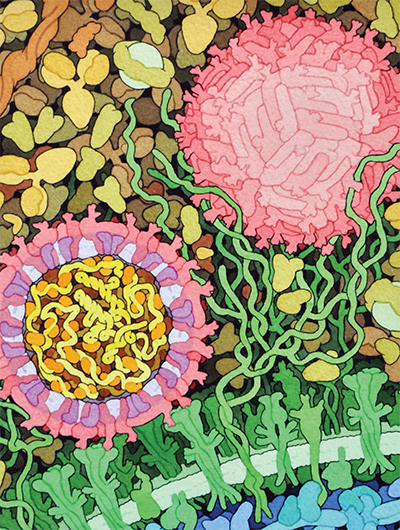 This space-fill drawing shows the outside of one Zika virus particle (red and pink) and a cross-section through another as it interacts with a cell. Courtesy of David Goodsell/Wikimedia Commons
This space-fill drawing shows the outside of one Zika virus particle (red and pink) and a cross-section through another as it interacts with a cell. Courtesy of David Goodsell/Wikimedia Commons
Like most viruses, Zika accomplishes a lot with a few tools. It has just one protein coding gene, which produces a single polypeptide that’s cleaved into 10 smaller proteins — a number dwarfed by the estimated 20,000 protein-coding genes in a human cell. Nevertheless, Zika can take over the vastly more complex human cell, repurposing it into a virus factory. Brian Raught, a researcher at the University of Toronto, said he found that process fascinating.
“With just these 10 proteins, this crazy virus turns your cells into zombies that do its bidding,” Raught said. “I always found that mind-blowing.”
Researchers in Raught’s lab, led by postdoctoral fellow Etienne Coyaud, wanted to find out how the handful of Zika proteins were able to hijack the host cell. They knew that the feat must depend on physical interactions between viral proteins and proteins native to the cell — but which ones?
Because of the increasing evidence linking Zika infections in expectant mothers to microcephaly in their children, Raught said, “We thought it better to leave the actual virus work to the experts.”
Instead of using infectious material, the team made 10 strains of human cells, each expressing one of Zika’s 10 proteins. By adding a small epitope tag to each viral protein, they were able to retrieve the viral proteins using an antibody that binds to this tag. The host proteins that stuck tightly to each viral protein came along for the ride; the researchers used mass spectrometry to identify those human proteins.
But there was a drawback to using this approach. What if proteins from the human cell were interacting with Zika proteins but separating from them before being extracted? To identify proteins that are close to but not inseparably intertwined with each viral protein, the team used a second technique called proximity labeling. In essence, they rigged each viral protein with an enzyme that would attach a sticky biotin tag onto anything that came close enough.
Proximity labeling is especially useful for detecting interactions with proteins embedded in cell membranes; those molecules are notoriously difficult to isolate.
“This is a really big asset of a proximity-labeling approach compared to traditional approaches,” Coyaud said.
Because Zika, like many viruses, enters the cell in a membrane-bound envelope and reorganizes many of its host’s membrane-bound organelles in the course of infection, its interactions with membrane proteins might be key to understanding the viral life cycle.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Bacterial enzyme catalyzes body odor compound formation
Researchers identify a skin-resident Staphylococcus hominis dipeptidase involved in creating sulfur-containing secretions. Read more about this recent Journal of Biological Chemistry paper.

Neurobiology of stress and substance use
MOSAIC scholar and proud Latino, Bryan Cruz of Scripps Research Institute studies the neurochemical origins of PTSD-related alcohol use using a multidisciplinary approach.

Pesticide disrupts neuronal potentiation
New research reveals how deltamethrin may disrupt brain development by altering the protein cargo of brain-derived extracellular vesicles. Read more about this recent Molecular & Cellular Proteomics article.

A look into the rice glycoproteome
Researchers mapped posttranslational modifications in Oryza sativa, revealing hundreds of alterations tied to key plant processes. Read more about this recent Molecular & Cellular Proteomics paper.

Proteomic variation in heart tissues
By tracking protein changes in stem cell–derived heart cells, researchers from Cedars-Sinai uncovered surprising diversity — including a potential new cell type — that could reshape how we study and treat heart disease.

Parsing plant pigment pathways
Erich Grotewold of Michigan State University, an ASBMB Breakthroughs speaker, discusses his work on the genetic regulation of flavonoid biosynthesis.

