Nuclear phosphoinositides: unruly little lipids
The biochemical problem presented by phosphoinositide lipids located in the cell nucleus is unique: These phospholipids exist outside nuclear membranes within the nucleoplasm, held soluble by largely unidentified nuclear proteins. Although endonuclear lipids have been observed in all eukaryotes, we still know next to nothing about what these powerful signaling molecules do in the nucleus, how they do it or why evolution chose this unorthodox physicochemical format.
The enigmatic history of nuclear phosphoinositides is well documented, reflecting how well these baffling biomolecules have evaded biochemical characterization. However, we were able to shed some light on the problem with a series of crystal structures of nuclear receptor transcription factors. These structures showed that certain nuclear receptors are able to solubilize phosphoinositide lipids by burying the acyl chains deep in the hydrophobic core of the protein, while the hydrophilic head groups remain solvent-exposed. We also found that particular lipid signaling enzymes can remodel the solvent-exposed head group with excellent kinetic properties, regulating the biological function of the nuclear receptors in transcriptional activation (illustration).
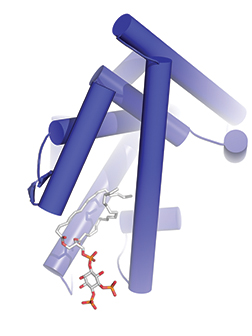 Phosphoinositide lipid acyl chains are buried in the hydrophobic core of the nuclear receptor, while the head group is highly exposed to solvent.Image courtesy of Ray Blind
Phosphoinositide lipid acyl chains are buried in the hydrophobic core of the nuclear receptor, while the head group is highly exposed to solvent.Image courtesy of Ray Blind
These discoveries presented a clear paradigm describing how endonuclear phosphoinositides can float about in the nucleoplasm and identified a specific biological activity they regulate.
However, many questions remain, and first on that list is the precise molecular event that is mediated by nuclear phosphoinositides. Further complicating the story is that the genes encoding the nuclear receptors we characterized exist only in metazoans, so the story in other members of eukaryota is wide open. Our group and Aurelia Lewis’ group at the University of Bergen, among others, are working feverishly to reveal the identities of these nuclear phospholipid effectors and determine how they trigger their nuclear effects.
Regardless of which nuclear proteins are regulated by nuclear phospholipids, the most intriguing question for me is why endonuclear lipids exist in the first place. Our work showed that lipid-signaling enzymes can phosphorylate phosphoinositides bound to nuclear proteins. But what inherent advantage would phosphorylating a signaling molecule tightly bound to a protein have over phosphorylating an amino acid in the protein itself?
Before sinking too deep in your thought experiments, let it be known that our nuclear receptors bind many different phospholipids with chemically unique head groups, each head group either compatible or incompatible with any given lipid signaling enzyme: Thus, by phosphorylating a dynamically exchangeable second messenger associated with an effector protein rather than a genomically encoded amino acid of the effector protein, the cell acquires the ability to decouple the effector protein from highly integrated signaling networks, disentangling particular circuits within large signaling architectures. We’re only starting to scratch the surface of the biological information-storage and -transfer implications stemming from this new, very intriguing hypothesis.
We now have many new tools in place to address longstanding questions in the nuclear lipid signaling field. The innovative and difficult work being done by those in this enigmatic field will clarify the nuclear roles of phosphoinositides, hopefully revealing new, undiscovered evolutionary design principles fundamental to biological signaling.
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Redefining lipid biology from droplets to ferroptosis
James Olzmann will receive the ASBMB Avanti Award in Lipids at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Women’s health cannot leave rare diseases behind
A physician living with lymphangioleiomyomatosis and a basic scientist explain why patient-driven, trial-ready research is essential to turning momentum into meaningful progress.

Life in four dimensions: When biology outpaces the brain
Nobel laureate Eric Betzig will discuss his research on information transfer in biology from proteins to organisms at the 2026 ASBMB Annual Meeting.

Fasting, fat and the molecular switches that keep us alive
Nutritional biochemist and JLR AE Sander Kersten has spent decades uncovering how the body adapts to fasting. His discoveries on lipid metabolism and gene regulation reveal how our ancient survival mechanisms may hold keys to modern metabolic health.

Redefining excellence to drive equity and innovation
Donita Brady will receive the ASBMB Ruth Kirschstein Award for Maximizing Access in Science at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

Mining microbes for rare earth solutions
Joseph Cotruvo, Jr., will receive the ASBMB Mildred Cohn Young Investigator Award at the ASBMB Annual Meeting, March 7–10, just outside of Washington, D.C.

