New tools to study protein–protein interactions in zebra fish
Understanding protein interactions and how proteins are organized in cellular compartments is essential to understanding their function. Since 2012, researchers have been using a technique called enzyme-catalyzed
In this technique, an enzyme, usually biotin ligase, also called BioID, is fused to a protein of interest, or bait protein, and exogenously introduced into cells. The cells then are supplemented with substrates that cause the biotin ligase to add biotin tags to other proteins in proximity of or interacting with the target protein. These biotinylated proteins then can be enriched, isolated and identified using liquid chromatography coupled with mass spectrometry.
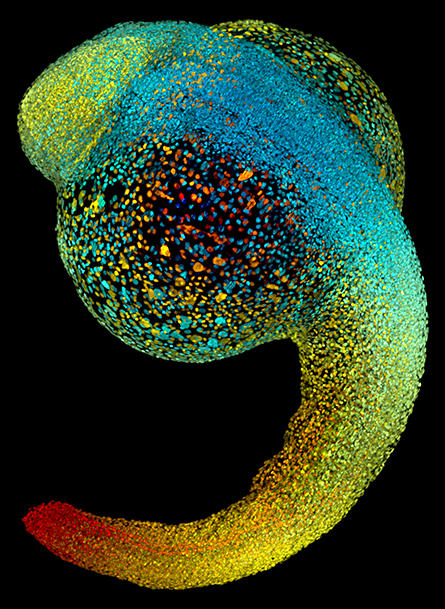
hours, all of the major organs will have started to form.
Anne-Claude Gingras’ research lab at the University of Toronto was one of the earliest adopters of enzyme-catalyzed proximity labeling.
“My lab has extensively used the BioID enzyme to label different proteins in living cells and report on their organization in cells and identify new components of organelles and protein binding partners,” Gingras said. “However, like many other researchers using proximity labeling, our studies have been limited to cells in culture.”
Studies assessing proximity labeling in animal models, including vertebrates such as zebra fish, have been limited.
Ian Scott is also a researcher at Toronto; his lab uses zebra fish to study early cardiac and vascular development. “Zebra fish embryos develop outside the mother’s body, and most of their organs appear within 36 hours of fertilization,” Scott said. “This makes them an ideal model for studying early development.”
As a first-year graduate student at the university, Shimon Rosenthal pursued research rotations in both Gingras’ and Scott’s labs. For his doctoral research, he was interested in using genetic and proteomics tools to study cardiac development in animal models.
“We were interested in establishing a proximity labeling approach in the zebra fish model,” Rosenthal said. “The project really interested me, and I welcomed the opportunity to be co-mentored by (Gingras and Scott).”
The project proved to be a challenge in the initial stages. It turned out that the widely used BioID enzyme was not effective for proximity labeling in zebra fish embryos. Eventually, two mutants of BioID — TurboID and miniTurbo — developed by Alice Ting’s research group at Stanford University demonstrated effective proximity labeling in zebra fish embryos.
Through meticulous experimentation, Rosenthal successfully developed and optimized TurboID and miniTurbo labeling in early zebra fish embryos. In addition, he demonstrated the versatility of his methodology through commonly used methods of exogenous protein introduction into cells. The methodology was recently published in a paper in the journal Molecular & Cellular Proteomics.
“We not only proved that our protocol works, but that it is just as effective as the proximity labeling technique in cultured cells,” Rosenthal said.
By generating transgenic zebra fish lines, Rosenthal was also able to study protein–protein interactions in specific tissues. Gingras and Scott will use this approach to check protein–protein interactions in diseases such as cerebral cavernous malformations, a disorder characterized by abnormal vessels in the brain’s vasculature that are prone to leak blood.
“In addition,” Scott said, “we hope to perform some experiments in mutant zebra fish models and understand how getting rid of one protein affects protein–protein interactions.”
Gingras and Scott have received positive feedback for their work, and both look forward to other researchers incorporating enzyme-catalyzed proximity labeling for their animal studies.
Enjoy reading ASBMB Today?
Become a member to receive the print edition monthly and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

Weedy rice gets competitive boost from its wild neighbors
Rice feeds the world. But researchers have found that a look-alike weed has many ways of getting ahead.

From the journals: JLR
A “T” makes a difference in blood clotting. High cholesterol: two screens are better than one. Biomarkers for cardiovascular risk. Statin-induced changes to the HDL lipidome. Read about recent papers on these topics.

Decoding microglial language
Emory University scientists characterize extracellular vesicles that facilitate intercellular communication.

What is metabolism?
A biochemist explains how different people convert energy differently – and why that matters for your health.

What’s next in the Ozempic era?
Diabetes, weight loss and now heart health: A new family of drugs is changing the way scientists are thinking about obesity — and more uses are on the horizon.
How a gene spurs tooth development
University of Iowa researchers find a clue in a rare genetic disorder’s missing chromosome.

