A chain of events
A few years ago, David Komander was tipped off to a mysterious gene. Komander, a biochemist at the Medical Research Council Laboratory of Molecular Biology in the U.K., is an expert in the enzymes of the ubiquitin system. A colleague told him that he had found a gene that made a protein of unknown function called FAM105B. “If any protein is called FAM,” says Komander, “it usually means nobody knows anything about it.”
The gene for FAM105B bore some of the hallmarks of those that make deubiquitinases. Deubiquitinases are enzymes that remove chains of ubiquitin. After doing their analyses, Komander and his colleagues concluded in 2013 that FAM105B was a deubiquitinase.
But it wasn’t just any deubiquitinase. It was specific to a special kind of ubiquitin chain. After finding this particular deubiquitinase, several groups of researchers, including Komander’s, discovered that the enzyme was important for keeping immune cells on an exquisitely delicate balance. If something went wrong with the enzyme, the immune cells went off the rails and caused a rare inflammatory disease.
 In the absence of functional OTULIN, defective immune cells can’t control signaling pathways regulated by linear ubiquitin chains. They send out signals that lead to a systemic inflammatory response. The hyper-active immune cells damage healthy tissues.
Courtesy of Lesley McKeane, MRC-LMB Visual Aids Department
In the absence of functional OTULIN, defective immune cells can’t control signaling pathways regulated by linear ubiquitin chains. They send out signals that lead to a systemic inflammatory response. The hyper-active immune cells damage healthy tissues.
Courtesy of Lesley McKeane, MRC-LMB Visual Aids Department
OTULIN outed
Ubiquitin is a protein of merely 76 amino acids. When tacked onto proteins, ubiquitin affects them in a number of ways. For one, it can set off the degradation of the tagged protein, a discovery that got Irwin Rose at the University of California, Irvine, as well as Aaron Ciechanover and Avram Hershko at the Technion–Israel Institute of Technology the Nobel Prize in chemistry in 2004. Ubiquitination also can change a protein’s location, activity or ability to interact with other proteins.
A triad of enzymes known as E1s, E2s and E3s attach ubiquitin to proteins. Ubiquitin can be attached either as a single molecule or as a chain. Chains of ubiquitin have a variety of forms, because the chains can be linked in eight different ways, either through one of the seven lysines or a specific methionine called Met1 on the ubiquitin molecule. Given the different ways for attaching ubiquitin — single or one of the eight types of chains — the repertoire of ubiquitin modifications is extensive. Different types of modifications have their own effects and sets of enzymes.
However, the existence of one of the ubiquitin chains, the linear chains made through Met1, once was contested. About five years ago, it was “hotly debated whether these linear ubiquitin chains are very important cellular signals,” says Komander. Researchers were unable to detect Met1-linked chains readily and they didn’t know of any enzyme responsible for removing the Met1-linked chains. Scientists had found the linear ubiquitin assembly complex, known as LUBAC, which built up the linear ubiquitin chains, but no one had found the enzymes that specifically removed them. “This was really the key missing activity,” says Komander. If a signal is important, there must be enzymes in place to put the signal on and take it off for the sake of regulation.
With the deubiquitinase for Met1-linked linear ubiquitin chains missing, some researchers were doubtful that Met1-linked linear ubiquitin chains were critical in cell physiology. But after analyzing FAM105B, Komander and others had their hands on the missing enzyme. It was the proof that linear chains are important in cell signaling.
In a 2013 Cell paper, Komander and his team described how their deubiquitinase specifically cleaved linear ubiquitin chains. Because the enzyme belonged to the OTU family of deubiquitinases and was targeted solely for linear ubiquitin chains, the investigators put OTU and “lin” together to give “OTULIN.” The same protein also was found by Frank Sicheri and Sabine Cordes at Samuel Lunenfeld Research Institute in Canada; they called the protein Gumby in their 2013 Nature paper.
Molecular biologists now had the deubiquitinase that was specific for breaking down linear ubiquitin chains. But clinicians soon became intrigued by the enzyme.
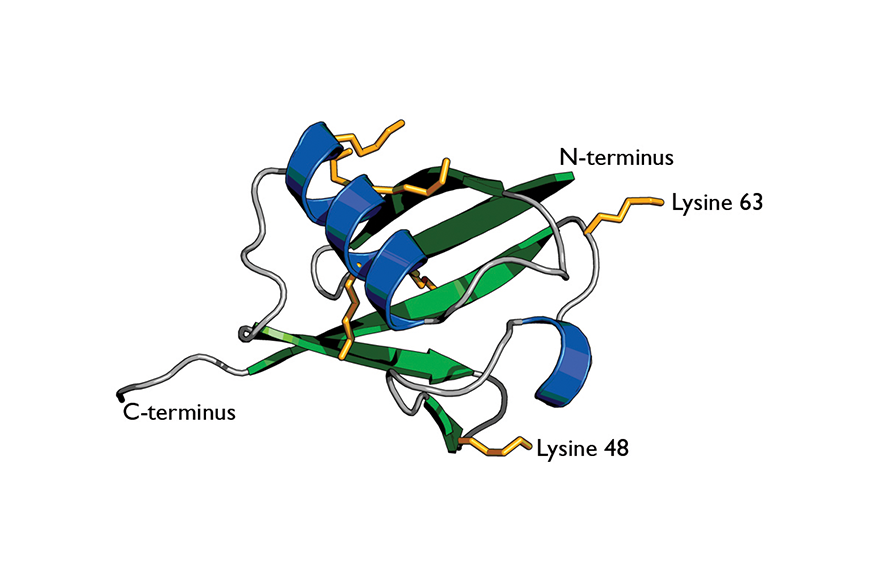 Cartoon of ubiquitin highlights its secondary structure.
Wikimedia
Cartoon of ubiquitin highlights its secondary structure.
Wikimedia
The gene hunt
Ivona Aksentijevich at the National Human Genome Research Institute has devoted her medical career to studying patients who have disorders that involve systemic inflammation, the causes for which are unknown. “I’ve been studying patients with rare inflammatory diseases for more than 25 years,” notes Aksentijevich. “I have participated in gene discoveries for nine diseases.”
So it wasn’t out of the ordinary that she and her colleague, Daniel Kastner, also at the NHGRI, were asked to examine a British Pakistani family with a child who displayed severe systemic inflammation from birth. The child was kept alive by heavy doses of anti-inflammatory medications and steroids. The child had two unaffected siblings but had first cousins who had symptoms of systemic inflammation at birth and died early in childhood.
The important clue clinicians had was that the family had intermarried for several generations. The disease was unknown outside of the family and not described in literature. Whatever was causing the disease had to be a genetic mistake that was carried in a single gene in both the parents’ DNA.
Aksentijevich, Kastner and the rest of their team collected blood samples from the family members and carried out whole-exome sequencing. They noted that the child with the rare disorder “had a homozygous mutation in this protein which is known as OTULIN.” Aksentijevich adds that the researchers first came across the name FAM105B, “which misled us for some time because we thought it was some poorly characterized gene.” But once they realized they were dealing with OTULIN, the clinicians knew what they were working with.
The investigators knew about patients who had a deficiency in LUBAC and suffered from an unusual phenotype that involved the immune system. OTULIN was known to interact with the LUBAC complex, which suggested that mutations in OTULIN also could cause systemic inflammation.
But, Aksentijevich says, “in studies where we define a new disease-causing gene, it makes for much stronger evidence to have additional patients with mutations in the same gene.” So she went to the database that she and her team had built up over the years of patients with mystery disorders and searched for those who had similar features to the British Pakistani patient they had analyzed. She found another patient from Turkey who carried a different homozygous mutation in OTULIN. And like that, Aksentijevich identified the second family with an OTULIN mutation.
Aksentijevich also reached out to collaborators in Turkey, a country where intermarriage is common, and asked if they knew of any other families that had the clinical symptoms of systemic inflammation and mutations in the OTULIN gene. Together, they identified a third family with a child who carried yet another type of mutation in OTULIN. This now meant “we had three different homozygous mutations in this gene,” says Aksentijevich. “That was a solid genetic evidence that OTULIN is a disease-causing gene.” Aksentijevich, Kastner and colleagues did experiments to show that these mutations cause the deficiency of OTULIN activity in the patients’ cells.
Later, during a conference in Europe, Aksentijevich heard about a Japanese girl who had similar clinical features but different mutations in OTULIN. So far, four patients have been identified with the OTULIN deficiency.
Across the Atlantic Ocean, Komander and his team also were concluding that homozygous recessive mutations in OTULIN caused inflammatory symptoms. Excited by the finding that OTULIN was the missing deubiquitinase for linear ubiquitin chains, Rune Damgaard, a postdoctoral fellow in Komander’s laboratory, began to develop knockout mice based on OTULIN in close collaboration with the group of Andrew McKenzie at the MRC Laboratory of Molecular Biology.
Researchers already knew that complete removal of OTULIN killed mice at the embryonic stage. When Damgaard selectively knocked out the gene in different cell types of the immune system, the mice developed signs of systemic inflammation. They had high levels of inflammatory mediators in their blood. Their white-blood cell count was high, as if the body were trying to fend off an imaginary infection. The overactive immune system was damaging the liver and other healthy organs.
Around this time, a clinician named Eamonn Maher got in touch with Komander because he had heard of the work going on in his laboratory. Maher had been treating the British Pakistani child in Birmingham, U.K., since the late 1990s with heavy doses of anti-inflammatory medications to keep the child alive. Maher had investigated the genes of this patient and the patient’s family and found mutations in FAM105B. But he had no idea what FAM105B did and couldn’t make sense of his finding. Then he learned about the Komander group’s work in identifying FAM105B as OTULIN.
Maher met with Komander and Damgaard over coffee and described his patient. Damgaard found himself nodding along as Maher listed the child’s symptoms. He was seeing similar symptoms in the OTULIN-knockout mice he was creating. Both the child and mice had overactive immune systems. The only difference was that the child had additional complications of large, patchy skin rashes and persistent diarrhea. Aksentijevich, Kastner and colleagues noted the same symptoms with their Turkish patients and independently had examined the genome of the British Pakistani family as had Maher.
When Komander and his colleagues published their paper in Cell in August, they called the disease ORAS for OTULIN-related autoinflammatory syndrome. Aksentijevich, Kastner and colleagues, in their paper, which appeared two weeks later in the Proceedings of the National Academy of Sciences, called the disease otulipenia. The two groups of investigators were aware of each other’s work and were excited to learn that their results, taken from cell lines, mice and patients (both groups had analyzed cells from the British Pakistani family), corroborated one another.
“We expect more patients to be diagnosed now that we know what to look for,” says Aksentijevich. She points out that identifying the disease and its cause helps physicians, because they will be able immediately to prescribe an anti-inflammatory treatment instead of first trying aggressive steroid treatments, which can cause major health complications.
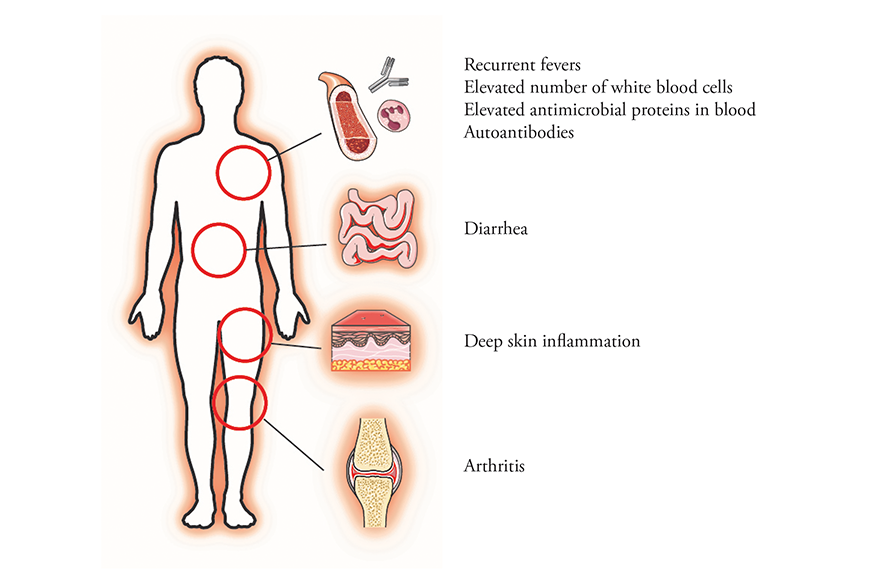 Mutations in the gene for OTULIN cause a rare systemic inflammatory disease.
Courtesy of Rune Damgaard and David Komander
Mutations in the gene for OTULIN cause a rare systemic inflammatory disease.
Courtesy of Rune Damgaard and David Komander
A fine balance
As investigators gather molecular and clinical information about OTULIN, they are marveling at the potency of linear ubiquitin chains. One of the critical pathways regulated by these chains is the NF-κB signaling pathway, which plays a key role in regulating the immune response to infection.
Under normal conditions, the linear ubiquitin chains mediate a time-dependent activation of NF-κB. The OTULIN mutations that have been found in patients all cause a severe reduction in the activity or the amount of the enzyme in cells compared with what normally is found in people. With lower levels of the enzyme, the linear ubiquitin chains don’t get chopped off, and this keeps the NF-κB signaling pathway firing without restrictions. At the physiological level, the immune system keeps chasing after an imaginary infection.
Scientists find it interesting that mutations in LUBAC also cause the immune system to go into overdrive and cause inflammatory disorders. It appears that the linear ubiquitin chain type is so highly regulated that, if the system goes awry in either way, the result is an overactive immune system.
“It just shows how exquisitely balanced this signal is in the cell,” says Komander. “For me, that is the most striking aspect. We’re talking here about a chain type that many people contested even to exist.”
Enjoy reading ASBMB Today?
Become a member to receive the print edition monthly and the digital edition weekly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles
Iron could be key to treating a global parasitic disease
A study has found that leishmaniasis causes body-wide changes in iron balance, leading to red blood cell damage.

Environmental DNA is everywhere
The ability to extract trace bits of DNA from soil, water, and even air is revolutionizing science. Are there pitfalls?
Early COVID-19 research is riddled with poor methods and low-quality results
The pandemic worsened, but didn’t create, this problem for science.

From the journals: MCP
Three views of mass spec: analyzing secreted protein spectra, imaging mass spectrometry for clinical use and spectral libraries for MS data analysis. Read about these recent papers.

Understanding the fat science
Researchers at UCLA investigate lipid remodeling in the liver for energy generation.

No oxygen? No problem
By studying how electric fish survive in hypoxic streams for months at time, researchers may find new ways to target tumors.

